3Q3C
 
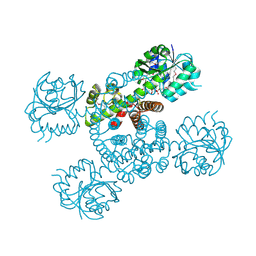 | | Crystal structure of a serine dehydrogenase from Pseudomonas aeruginosa pao1 in complex with NAD | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Probable 3-hydroxyisobutyrate dehydrogenase | | Authors: | Tan, K, Singer, A.U, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A.M, Joachimiak, A, Yakunin, A.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-12-21 | | Release date: | 2011-02-23 | | Last modified: | 2012-02-01 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | Biochemical and Structural Studies of Uncharacterized Protein PA0743 from Pseudomonas aeruginosa Revealed NAD+-dependent L-Serine Dehydrogenase.
J.Biol.Chem., 287, 2012
|
|
3Q13
 
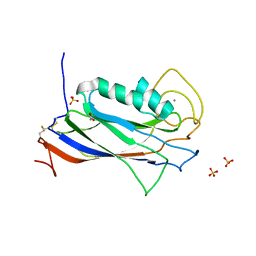 | | The Structure of the Ca2+-binding, Glycosylated F-spondin Domain of F-spondin, A C2-domain Variant from Extracellular Matrix | | Descriptor: | ACETATE ION, CALCIUM ION, SULFATE ION, ... | | Authors: | Tan, K, Lawler, J. | | Deposit date: | 2010-12-16 | | Release date: | 2011-06-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The structure of the Ca2+-binding, glycosylated F-spondin domain of F-spondin - A C2-domain variant in an extracellular matrix protein.
Bmc Struct.Biol., 11, 2011
|
|
2IA9
 
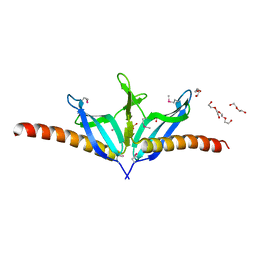 | | Structural Genomics, the crystal structure of SpoVG from Bacillus subtilis subsp. subtilis str. 168 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Putative septation protein spoVG, SULFATE ION | | Authors: | Tan, K, Borovilos, M, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-09-07 | | Release date: | 2006-10-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The crystal structure of SpoVG from Bacillus subtilis subsp. subtilis str. 168
To be Published
|
|
6W4B
 
 | | The crystal structure of Nsp9 RNA binding protein of SARS CoV-2 | | Descriptor: | Non-structural protein 9 | | Authors: | Tan, K, Kim, Y, Jedrzejczak, R, Maltseva, N, Endres, M, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-03-10 | | Release date: | 2020-03-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | The crystal structure of Nsp9 replicase protein of COVID-19
To Be Published
|
|
2I5E
 
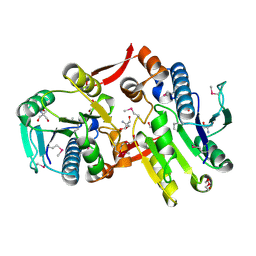 | | Crystal Structure of a Protein of Unknown Function MM2497 from Methanosarcina mazei Go1, Probable Nucleotidyltransferase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, Hypothetical protein MM_2497 | | Authors: | Tan, K, Du, J, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-08-24 | | Release date: | 2006-09-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of a hypothetical protein MM_2497 from Methanosarcina mazei Go1
To be Published
|
|
1L6Z
 
 | | CRYSTAL STRUCTURE OF MURINE CEACAM1A[1,4]: A CORONAVIRUS RECEPTOR AND CELL ADHESION MOLECULE IN THE CEA FAMILY | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, biliary glycoprotein C | | Authors: | Tan, K, Zelus, B.D, Meijers, R, Liu, J.-H, Bergelson, J.M, Duke, N, Zhang, R, Joachimiak, A, Holmes, K.V, Wang, J.-H. | | Deposit date: | 2002-03-14 | | Release date: | 2002-09-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.32 Å) | | Cite: | CRYSTAL STRUCTURE OF MURINE sCEACAM1a[1,4]: A CORONAVIRUS RECEPTOR IN THE CEA FAMILY
Embo J., 21, 2002
|
|
2HKU
 
 | | Structural Genomics, the crystal structure of a putative transcriptional regulator from Rhodococcus sp. RHA1 | | Descriptor: | 1,2-ETHANEDIOL, A putative transcriptional regulator, TETRAETHYLENE GLYCOL | | Authors: | Tan, K, Evdokimova, E, Kagan, O, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-07-05 | | Release date: | 2006-08-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of a transcriptional regulator from Rhodococcus sp. RHA1
To be Published
|
|
3DF8
 
 | |
3D8U
 
 | |
3FC7
 
 | |
3FK8
 
 | |
3F1B
 
 | | The crystal structure of a TetR-like transcriptional regulator from Rhodococcus sp. RHA1. | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, TetR-like transcriptional regulator | | Authors: | Tan, K, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-10-27 | | Release date: | 2008-11-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of a TetR-like transcriptional regulator from Rhodococcus sp. RHA1.
To be Published
|
|
6V6N
 
 | | The crystal structure of a class D beta-lactamase from Agrobacterium tumefaciens | | Descriptor: | Beta-lactamase, FORMIC ACID, GLYCEROL, ... | | Authors: | Tan, K, Wu, R, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-12-05 | | Release date: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The crystal structure of a class D beta-lactamase from Agrobacterium tumefaciens
To Be Published
|
|
3DSB
 
 | |
6V4W
 
 | | The crystal structure of a beta-lactamase from Chitinophaga pinensis DSM 2588 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ACETATE ION, Beta-lactamase, ... | | Authors: | Tan, K, Welk, L, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-12-02 | | Release date: | 2019-12-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | The crystal structure of a beta-lactamase from Chitinophaga pinensis DSM 2588
To Be Published
|
|
3E0X
 
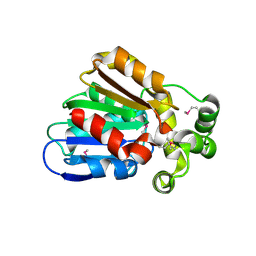 | |
3F5B
 
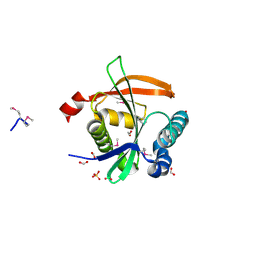 | | The crystal structure of aminoglycoside N(6')acetyltransferase from Legionella pneumophila subsp. pneumophila str. Philadelphia 1. | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Aminoglycoside N(6')acetyltransferase, ... | | Authors: | Tan, K, Wu, R, Perez, V, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-11-03 | | Release date: | 2008-11-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of aminoglycoside N(6')acetyltransferase from Legionella pneumophila subsp. pneumophila str. Philadelphia 1.
To be Published
|
|
3F5R
 
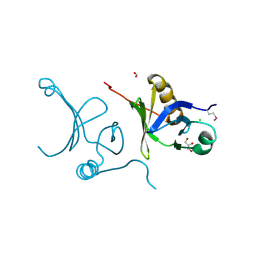 | | The crystal structure of a subunit of the heterodimeric FACT complex (Spt16p-Pob3p). | | Descriptor: | CHLORIDE ION, FACT complex subunit POB3, FORMIC ACID, ... | | Authors: | Tan, K, Xu, X, Cui, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-11-04 | | Release date: | 2008-11-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structure of a subunit of the heterodimeric FACT complex (Spt16p-Pob3p).
To be Published
|
|
3COO
 
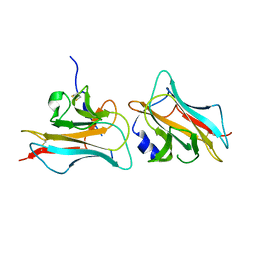 | |
3CRJ
 
 | |
3E0Y
 
 | |
3FZ4
 
 | | The crystal structure of a possible arsenate reductase from Streptococcus mutans UA159 | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, Putative arsenate reductase, ... | | Authors: | Tan, K, Hatzos, C, Shackelford, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-01-23 | | Release date: | 2009-02-10 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | The crystal structure of a possible arsenate reductase from Streptococcus mutans UA159.
To be Published
|
|
6WGQ
 
 | | The crystal structure of a beta-lactamase from Shigella flexneri 2a str. 2457T | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-lactamase, CHLORIDE ION, ... | | Authors: | Tan, K, Wu, R, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-04-06 | | Release date: | 2020-04-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of a beta-lactamase from Shigella flexneri 2a str. 2457T
To Be Published
|
|
3FQ6
 
 | |
3DNH
 
 | | The crystal structure of the protein Atu2129 (unknown function) from Agrobacterium tumefaciens str. C58 | | Descriptor: | uncharacterized protein Atu2129 | | Authors: | Tan, K, Xu, X, Zheng, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-07-02 | | Release date: | 2008-09-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | The crystal structure of the protein Atu2129 (unknown function) from Agrobacterium tumefaciens str. C58
To be Published
|
|
