2OYW
 
 | | Neurotensin in TFE:H2O (80:20) | | Descriptor: | neurotensin | | Authors: | Monti, J.P, Coutant, J, Curmi, P.A. | | Deposit date: | 2007-02-23 | | Release date: | 2007-05-08 | | Last modified: | 2019-12-25 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of Neurotensin in Membrane-Mimetic Environments: Molecular Basis for Neurotensin Receptor Recognition.
Biochemistry, 46, 2007
|
|
2M3W
 
 | |
1GNM
 
 | | HIV-1 PROTEASE MUTANT WITH VAL 82 REPLACED BY ASP (V82D) COMPLEXED WITH U89360E (INHIBITOR) | | Descriptor: | HIV-1 PROTEASE, N-[[1-[N-ACETAMIDYL]-[1-CYCLOHEXYLMETHYL-2-HYDROXY-4-ISOPROPYL]-BUT-4-YL]-CARBONYL]-GLUTAMINYL-ARGINYL-AMIDE | | Authors: | Hong, L, Treharne, A, Hartsuck, J.A, Foundling, S, Tang, J. | | Deposit date: | 1996-05-04 | | Release date: | 1996-11-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of complexes of a peptidic inhibitor with wild-type and two mutant HIV-1 proteases.
Biochemistry, 35, 1996
|
|
4F1S
 
 | | Crystal structure of human PI3K-gamma in complex with a pyridyl-triazine-sulfonamide inhibitor | | Descriptor: | N-(5-{[3-(4-amino-6-methyl-1,3,5-triazin-2-yl)-5-(tetrahydro-2H-pyran-4-yl)pyridin-2-yl]amino}-2-chloropyridin-3-yl)methanesulfonamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | Authors: | Whittington, D.A, Tang, J, Yakowec, P. | | Deposit date: | 2012-05-07 | | Release date: | 2012-08-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Synthesis and structure-activity relationships of dual PI3K/mTOR inhibitors based on a 4-amino-6-methyl-1,3,5-triazine sulfonamide scaffold.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
1GNO
 
 | | HIV-1 PROTEASE (WILD TYPE) COMPLEXED WITH U89360E (INHIBITOR) | | Descriptor: | HIV-1 PROTEASE, N-[[1-[N-ACETAMIDYL]-[1-CYCLOHEXYLMETHYL-2-HYDROXY-4-ISOPROPYL]-BUT-4-YL]-CARBONYL]-GLUTAMINYL-ARGINYL-AMIDE | | Authors: | Hong, L, Treharne, A, Hartsuck, J.A, Foundling, S, Tang, J. | | Deposit date: | 1996-05-04 | | Release date: | 1996-11-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of complexes of a peptidic inhibitor with wild-type and two mutant HIV-1 proteases.
Biochemistry, 35, 1996
|
|
1D61
 
 | | THE STRUCTURE OF THE B-DNA DECAMER C-C-A-A-C-I-T-T-G-G: MONOCLINIC FORM | | Descriptor: | CACODYLATE ION, CALCIUM ION, DNA (5'-D(*CP*CP*AP*AP*CP*IP*TP*TP*GP*G)-3') | | Authors: | Lipanov, A, Kopka, M.L, Kaczor-Grzeskowiak, M, Quintana, J, Dickerson, R.E. | | Deposit date: | 1992-02-26 | | Release date: | 1993-04-15 | | Last modified: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure of the B-DNA decamer C-C-A-A-C-I-T-T-G-G in two different space groups: conformational flexibility of B-DNA.
Biochemistry, 32, 1993
|
|
1DA3
 
 | | THE CRYSTAL STRUCTURE OF THE TRIGONAL DECAMER C-G-A-T-C-G-6MEA-T-C-G: A B-DNA HELIX WITH 10.6 BASE-PAIRS PER TURN | | Descriptor: | CHLORIDE ION, DNA (5'-D(*CP*GP*AP*TP*CP*GP*(6MA)P*TP*CP*G)-3'), MAGNESIUM ION | | Authors: | Baikalov, I, Grzeskowiak, K, Yanagi, K, Quintana, J, Dickerson, R.E. | | Deposit date: | 1992-11-09 | | Release date: | 1993-04-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of the trigonal decamer C-G-A-T-C-G-6meA-T-C-G: a B-DNA helix with 10.6 base-pairs per turn.
J.Mol.Biol., 231, 1993
|
|
1GNN
 
 | | HIV-1 PROTEASE MUTANT WITH VAL 82 REPLACED BY ASN (V82N) COMPLEXED WITH U89360E (INHIBITOR) | | Descriptor: | HIV-1 PROTEASE, N-[[1-[N-ACETAMIDYL]-[1-CYCLOHEXYLMETHYL-2-HYDROXY-4-ISOPROPYL]-BUT-4-YL]-CARBONYL]-GLUTAMINYL-ARGINYL-AMIDE | | Authors: | Hong, L, Treharne, A, Hartsuck, J.A, Foundling, S, Tang, J. | | Deposit date: | 1996-05-04 | | Release date: | 1996-11-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of complexes of a peptidic inhibitor with wild-type and two mutant HIV-1 proteases.
Biochemistry, 35, 1996
|
|
4NBY
 
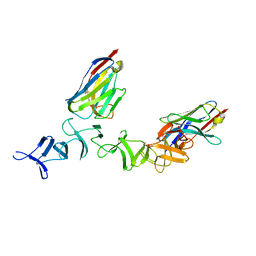 | | Crystal Structure of TcdA-A2 Bound to Two Molecules of A20.1 VHH | | Descriptor: | A20.1 VHH, Cell wall-binding repeat protein | | Authors: | Murase, T, Eugenio, L, Schorr, M, Hussack, G, Tanha, J, Kitova, E, Klassen, J.S, Ng, K.K.S. | | Deposit date: | 2013-10-23 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural Basis for Antibody Recognition in the Receptor-binding Domains of Toxins A and B from Clostridium difficile.
J.Biol.Chem., 289, 2014
|
|
4NC1
 
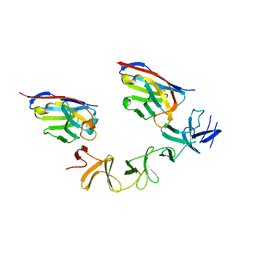 | | Crystal Structure of TcdA-A2 Bound to A20.1 VHH and A26.8 VHH | | Descriptor: | A20.1 VHH, A26.8 VHH, Cell wall-binding repeat protein | | Authors: | Murase, T, Eugenio, L, Schorr, M, Hussack, G, Tanha, J, Kitova, E.N, Klassen, J.S, Ng, K.K.S. | | Deposit date: | 2013-10-23 | | Release date: | 2013-12-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural Basis for Antibody Recognition in the Receptor-binding Domains of Toxins A and B from Clostridium difficile.
J.Biol.Chem., 289, 2014
|
|
4NC2
 
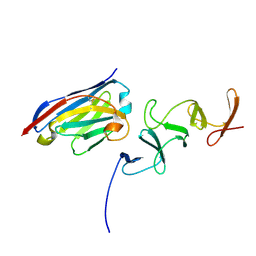 | | Crystal structure of TcdB-B1 bound to B39 VHH | | Descriptor: | B39 VHH, Toxin B | | Authors: | Murase, T, Eugenio, L, Schorr, M, Hussack, G, Tanha, J, Kitova, E.N, Klassen, J.S, Ng, K.K.S. | | Deposit date: | 2013-10-23 | | Release date: | 2013-12-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for Antibody Recognition in the Receptor-binding Domains of Toxins A and B from Clostridium difficile.
J.Biol.Chem., 289, 2014
|
|
4Q58
 
 | |
4Q59
 
 | |
4NBX
 
 | | Crystal Structure of Clostridium difficile Toxin A fragment TcdA-A1 Bound to A20.1 VHH | | Descriptor: | A20.1 VHH, TcdA | | Authors: | Murase, T, Eugenio, L, Schorr, M, Hussack, G, Tanha, J, Kitova, E.N, Klassen, J.S, Ng, K.K.S. | | Deposit date: | 2013-10-23 | | Release date: | 2013-12-11 | | Last modified: | 2014-02-12 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis for Antibody Recognition in the Receptor-binding Domains of Toxins A and B from Clostridium difficile.
J.Biol.Chem., 289, 2014
|
|
4NC0
 
 | | Crystal Structure of TcdA-A2 Bound to A26.8 VHH | | Descriptor: | A26.8 VHH, Cell wall-binding repeat protein | | Authors: | Murase, T, Eugenio, L, Schorr, M, Hussack, G, Tanha, J, Kitova, E.N, Klassen, J.S, Ng, K.K.S. | | Deposit date: | 2013-10-23 | | Release date: | 2013-12-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for Antibody Recognition in the Receptor-binding Domains of Toxins A and B from Clostridium difficile.
J.Biol.Chem., 289, 2014
|
|
3W6H
 
 | | Crystal structure of 19F probe-labeled hCAI in complex with acetazolamide | | Descriptor: | 1-(2-ethoxyethoxy)-3,5-bis(trifluoromethyl)benzene, 5-ACETAMIDO-1,3,4-THIADIAZOLE-2-SULFONAMIDE, Carbonic anhydrase 1, ... | | Authors: | Takaoka, Y, Kioi, Y, Morito, A, Otani, J, Arita, K, Ashihara, E, Ariyoshi, M, Tochio, H, Shirakawa, M, Hamachi, I. | | Deposit date: | 2013-02-14 | | Release date: | 2013-03-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.964 Å) | | Cite: | Quantitative Comparison of Protein Dynamics in Live Cells and In Vitro by In-Cell 19F-NMR
To be published
|
|
4Q57
 
 | | Crystal structure of the plectin 1a actin-binding domain/N-terminal domain of calmodulin complex | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Song, J.-G, Kostan, J, Grishkovskaya, I, Djinovic-Carugo, K. | | Deposit date: | 2014-04-16 | | Release date: | 2014-07-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the plectin 1a actin-binding domain/N-terminal domain of calmodulin complex
To be Published
|
|
3W6I
 
 | | Crystal structure of 19F probe-labeled hCAI | | Descriptor: | 1-(2-ethoxyethoxy)-3,5-bis(trifluoromethyl)benzene, Carbonic anhydrase 1, ZINC ION | | Authors: | Takaoka, Y, Kioi, Y, Morito, A, Otani, J, Arita, K, Ashihara, E, Ariyoshi, M, Tochio, H, Shirakawa, M, Hamachi, I. | | Deposit date: | 2013-02-14 | | Release date: | 2013-03-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.693 Å) | | Cite: | Quantitative Comparison of Protein Dynamics in Live Cells and In Vitro by In-Cell 19F-NMR
To be published
|
|
4NBZ
 
 | | Crystal Structure of TcdA-A1 Bound to A26.8 VHH | | Descriptor: | A26.8 VHH, TcdA | | Authors: | Murase, T, Eugenio, L, Schorr, M, Hussack, G, Tanha, J, Kitova, E.N, Klassen, J.S, Ng, K.K.S. | | Deposit date: | 2013-10-23 | | Release date: | 2013-12-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis for Antibody Recognition in the Receptor-binding Domains of Toxins A and B from Clostridium difficile.
J.Biol.Chem., 289, 2014
|
|
6HYK
 
 | | NMR solution structure of the C/D box snoRNA U14 | | Descriptor: | RNA (31-MER) | | Authors: | Chagot, M.E, Quinternet, M, Rothe, B, Charpentier, B, Coutant, J, Manival, X, Lebars, I. | | Deposit date: | 2018-10-22 | | Release date: | 2019-04-24 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The yeast C/D box snoRNA U14 adopts a "weak" K-turn like conformation recognized by the Snu13 core protein in solution.
Biochimie, 164, 2019
|
|
