2ZA8
 
 | | recombinant horse L-chain apoferritin N-terminal deletion mutant (residues 1-8) | | Descriptor: | CADMIUM ION, Ferritin light chain | | Authors: | Yamashita, I, Mishima, Y, Park, S.-Y, Heddle, J.G, Tame, J.R.H. | | Deposit date: | 2007-10-02 | | Release date: | 2008-01-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Effect of N-terminal Residues on the Structural Stability of Recombinant Horse L-chain Apoferritin in an Acidic Environment
J.BIOCHEM.(TOKYO), 142, 2007
|
|
2ZNL
 
 | | Crystal structure of PA-PB1 complex form influenza virus RNA polymerase | | Descriptor: | Polymerase acidic protein, RNA-directed RNA polymerase catalytic subunit | | Authors: | Obayashi, E, Yoshida, H, Kawai, F, Shibayama, N, Kawaguchi, A, Nagata, K, Tame, J.R.H, Park, S.-Y. | | Deposit date: | 2008-04-28 | | Release date: | 2008-09-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The structural basis for an essential subunit interaction in influenza virus RNA polymerase
Nature, 454, 2008
|
|
3VRE
 
 | | The crystal structure of hemoglobin from woolly mammoth in the deoxy form | | Descriptor: | Hemoglobin subunit alpha, Hemoglobin subunit beta/delta hybrid, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Noguchi, H, Campbell, K.L, Ho, C, Park, S.-Y, Tame, J.R.H. | | Deposit date: | 2012-04-09 | | Release date: | 2012-11-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of haemoglobin from woolly mammoth in liganded and unliganded states.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3VRG
 
 | | The crystal structure of hemoglobin from woolly mammoth in the met form | | Descriptor: | Hemoglobin subunit alpha, Hemoglobin subunit beta/delta hybrid, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Noguchi, H, Campbell, K.L, Ho, C, Park, S.-Y, Tame, J.R.H. | | Deposit date: | 2012-04-09 | | Release date: | 2012-11-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structures of haemoglobin from woolly mammoth in liganded and unliganded states.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3A3F
 
 | | Crystal structure of penicillin binding protein 4 (dacB) from Haemophilus influenzae,complexed with novel beta-lactam (FMZ) | | Descriptor: | (2R,4S)-5,5-dimethyl-2-[(1R)-2-oxo-1-({(2R)-2-[(2-oxoimidazolidin-1-yl)amino]-2-phenylacetyl}amino)ethyl]-1,3-thiazolidine-4-carboxylic acid, Penicillin-binding protein 4 | | Authors: | Kawai, F, Roper, D.I, Park, S.-Y, Tame, J.R.H. | | Deposit date: | 2009-06-12 | | Release date: | 2009-12-22 | | Last modified: | 2013-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of penicillin-binding proteins 4 and 5 from Haemophilus influenzae
J.Mol.Biol., 396, 2010
|
|
3A3I
 
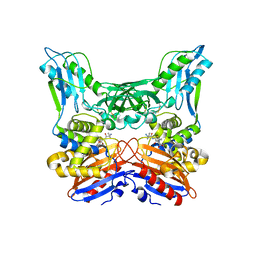 | | Crystal structure of penicillin binding protein 4 (dacB) from Haemophilus influenzae, complexed with ampicillin (AIX) | | Descriptor: | (2R,4S)-2-[(1R)-1-{[(2R)-2-amino-2-phenylacetyl]amino}-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, Penicillin-binding protein 4 | | Authors: | Kawai, F, Roper, D.I, Park, S.-Y, Tame, J.R.H. | | Deposit date: | 2009-06-12 | | Release date: | 2009-12-22 | | Last modified: | 2013-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of penicillin-binding proteins 4 and 5 from Haemophilus influenzae
J.Mol.Biol., 396, 2010
|
|
3WWF
 
 | | Crystal structure of the computationally designed Pizza2-SR protein | | Descriptor: | pizza2sr-pb protein | | Authors: | Voet, A.R.D, Noguchi, H, Addy, C, Simoncini, D, Terada, D, Unzai, S, Park, S.Y, Zhang, K.Y.J, Tame, J.R.H. | | Deposit date: | 2014-06-17 | | Release date: | 2014-10-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Computational design of a self-assembling symmetrical beta-propeller protein.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3WWB
 
 | | Crystal structure of the computationally designed Pizza2-SR protein | | Descriptor: | Pizza2-SR protein | | Authors: | Voet, A.R.D, Noguchi, H, Addy, C, Simoncini, D, Terada, D, Unzai, S, Park, S.Y, Zhang, K.Y.J, Tame, J.R.H. | | Deposit date: | 2014-06-17 | | Release date: | 2014-10-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Computational design of a self-assembling symmetrical beta-propeller protein.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3WMV
 
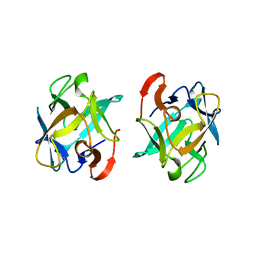 | | The structure of an anti-cancer lectin mytilec with ligand from the mussel Mytilus galloprovincialis | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, Lectin | | Authors: | Terada, D, Kawai, F, Noguchi, H, Unzai, S, Park, S.-Y, Ozeki, Y, Tame, J.R.H. | | Deposit date: | 2013-11-27 | | Release date: | 2014-12-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Crystal structure of MytiLec, a galactose-binding lectin from the mussel Mytilus galloprovincialis with cytotoxicity against certain cancer cell types
Sci Rep, 6, 2016
|
|
3WWA
 
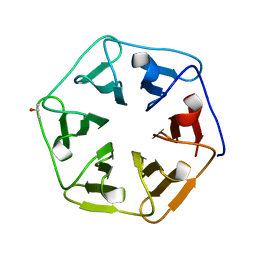 | | Crystal structure of the computationally designed Pizza7 protein after heat treatment | | Descriptor: | ISOPROPYL ALCOHOL, Pizza7H protein | | Authors: | Voet, A.R.D, Noguchi, H, Addy, C, Simoncini, D, Terada, D, Unzai, S, Park, S.Y, Zhang, K.Y.J, Tame, J.R.H. | | Deposit date: | 2014-06-17 | | Release date: | 2014-10-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.988 Å) | | Cite: | Computational design of a self-assembling symmetrical beta-propeller protein.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3WW9
 
 | | Crystal structure of the computationally designed Pizza6 protein | | Descriptor: | GLYCEROL, SULFATE ION, pizza6 protein | | Authors: | Voet, A.R.D, Noguchi, H, Addy, C, Simoncini, D, Terada, D, Unzai, S, Park, S.Y, Zhang, K.Y.J, Tame, J.R.H. | | Deposit date: | 2014-06-17 | | Release date: | 2014-10-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Computational design of a self-assembling symmetrical beta-propeller protein.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3WW7
 
 | | Crystal structure of the computationally designed Pizza2 protein | | Descriptor: | GLYCEROL, Pizza2 protein | | Authors: | Voet, A.R.D, Noguchi, H, Addy, C, Simoncini, D, Terada, D, Unzai, S, Park, S.Y, Zhang, K.Y.J, Tame, J.R.H. | | Deposit date: | 2014-06-17 | | Release date: | 2014-10-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.697 Å) | | Cite: | Computational design of a self-assembling symmetrical beta-propeller protein.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1B6H
 
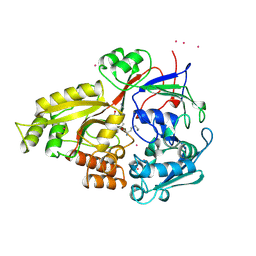 | |
1B7H
 
 | |
1B3H
 
 | |
1B2H
 
 | |
1B1H
 
 | | OLIGO-PEPTIDE BINDING PROTEIN/TRIPEPTIDE (LYS HPE LYS) COMPLEX | | Descriptor: | PROTEIN (LYS HPE LYS), PROTEIN (OLIGO-PEPTIDE BINDING PROTEIN), URANYL (VI) ION | | Authors: | Davies, T.G, Tame, J.R.H. | | Deposit date: | 1998-11-10 | | Release date: | 1998-11-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Relating structure to thermodynamics: the crystal structures and binding affinity of eight OppA-peptide complexes.
Protein Sci., 8, 1999
|
|
1B5H
 
 | |
1B4H
 
 | |
2EB4
 
 | | Crystal structure of apo-HpcG | | Descriptor: | 2-oxo-hept-3-ene-1,7-dioate hydratase, SODIUM ION, THIOCYANATE ION | | Authors: | Izumi, A, Rea, D, Adachi, T, Unzai, S, Park, S.Y, Roper, D.I, Tame, J.R.H. | | Deposit date: | 2007-02-07 | | Release date: | 2007-07-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and Mechanism of HpcG, a Hydratase in the Homoprotocatechuate Degradation Pathway of Escherichia coli
J.Mol.Biol., 370, 2007
|
|
2EB5
 
 | | Crystal structure of HpcG complexed with oxalate | | Descriptor: | 2-oxo-hept-3-ene-1,7-dioate hydratase, MAGNESIUM ION, OXALATE ION, ... | | Authors: | Izumi, A, Rea, D, Adachi, T, Unzai, S, Park, S.Y, Roper, D.I, Tame, J.R.H. | | Deposit date: | 2007-02-07 | | Release date: | 2007-07-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and Mechanism of HpcG, a Hydratase in the Homoprotocatechuate Degradation Pathway of Escherichia coli
J.Mol.Biol., 370, 2007
|
|
2EB6
 
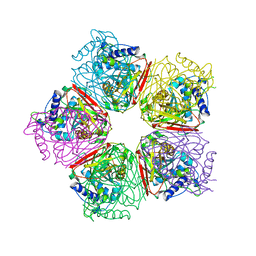 | | Crystal structure of HpcG complexed with Mg ion | | Descriptor: | 2-oxo-hept-3-ene-1,7-dioate hydratase, MAGNESIUM ION | | Authors: | Izumi, A, Rea, D, Adachi, T, Unzai, S, Park, S.Y, Roper, D.I, Tame, J.R.H. | | Deposit date: | 2007-02-07 | | Release date: | 2007-07-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structure and Mechanism of HpcG, a Hydratase in the Homoprotocatechuate Degradation Pathway of Escherichia coli
J.Mol.Biol., 370, 2007
|
|
2EX2
 
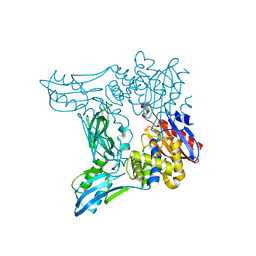 | | Crystal structure of penicillin binding protein 4 (dacB) from Escherichia coli | | Descriptor: | GLYCEROL, Penicillin-binding protein 4 | | Authors: | Kishida, H, Unzai, S, Roper, D.I, Lloyd, A, Park, S.-Y, Tame, J.R.H. | | Deposit date: | 2005-11-07 | | Release date: | 2006-06-13 | | Last modified: | 2016-10-19 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of penicillin binding protein 4 (dacB) from Escherichia coli, both in the native form and covalently linked to various antibiotics
Biochemistry, 45, 2006
|
|
2EXS
 
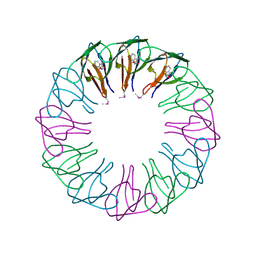 | | TRAP3 (engineered TRAP) | | Descriptor: | TRYPTOPHAN, Transcription attenuation protein mtrB | | Authors: | Heddle, J.G, Yokoyama, T, Yamashita, I, Park, S.Y, Tame, J.R.H. | | Deposit date: | 2005-11-08 | | Release date: | 2006-08-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Rounding up: Engineering 12-Membered Rings from the Cyclic 11-Mer TRAP
Structure, 14, 2006
|
|
2EXA
 
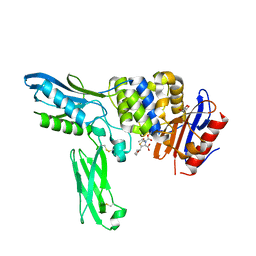 | | Crystal structure of penicillin binding protein 4 (dacB) from Escherichia coli, complexed with FAROM | | Descriptor: | (2R,5R)-2-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-5-[(2R)-tetrahydrofuran-2-yl]-2,5-dihydro-1,3-thiazole-4-carboxylic acid, GLYCEROL, Penicillin-binding protein 4 | | Authors: | Kishida, H, Unzai, S, Roper, D.I, Lloyd, A, Park, S.-Y, Tame, J.R.H. | | Deposit date: | 2005-11-08 | | Release date: | 2006-06-13 | | Last modified: | 2016-10-19 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of penicillin binding protein 4 (dacB) from Escherichia coli, both in the native form and covalently linked to various antibiotics
Biochemistry, 45, 2006
|
|
