7XE7
 
 | | T4 lysozyme mutant-S44C/C54T/N68C/A93C/C97A/T115C, pH10 | | Descriptor: | Endolysin, GLYCEROL, HEXANE-1,6-DIOL | | Authors: | Tamada, T, Hiromoto, T. | | Deposit date: | 2022-03-30 | | Release date: | 2023-03-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Creation of Cross-Linked Crystals With Intermolecular Disulfide Bonds Connecting Symmetry-Related Molecules Allows Retention of Tertiary Structure in Different Solvent Conditions.
Front Mol Biosci, 9, 2022
|
|
7XE6
 
 | | T4 lysozyme mutant-S44C/C54T/N68C/A93C/C97A/T115C, pH7 | | Descriptor: | Endolysin, GLYCEROL, HEXANE-1,6-DIOL, ... | | Authors: | Tamada, T, Hiromoto, T. | | Deposit date: | 2022-03-30 | | Release date: | 2023-03-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Creation of Cross-Linked Crystals With Intermolecular Disulfide Bonds Connecting Symmetry-Related Molecules Allows Retention of Tertiary Structure in Different Solvent Conditions.
Front Mol Biosci, 9, 2022
|
|
7XEA
 
 | | T4 lysozyme mutant-S44C/C54T/N68C/A93C/C97A/T115C, DMSO 40%, and then backsoaking | | Descriptor: | CHLORIDE ION, Endolysin, GLYCEROL, ... | | Authors: | Tamada, T, Hiromoto, T. | | Deposit date: | 2022-03-30 | | Release date: | 2023-03-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Creation of Cross-Linked Crystals With Intermolecular Disulfide Bonds Connecting Symmetry-Related Molecules Allows Retention of Tertiary Structure in Different Solvent Conditions.
Front Mol Biosci, 9, 2022
|
|
7XE9
 
 | | T4 lysozyme mutant-S44C/C54T/N68C/A93C/C97A/T115C, DMSO 20% | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, Endolysin, ... | | Authors: | Tamada, T, Hiromoto, T. | | Deposit date: | 2022-03-30 | | Release date: | 2023-03-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Creation of Cross-Linked Crystals With Intermolecular Disulfide Bonds Connecting Symmetry-Related Molecules Allows Retention of Tertiary Structure in Different Solvent Conditions.
Front Mol Biosci, 9, 2022
|
|
7XE5
 
 | | T4 lysozyme mutant-S44C/C54T/N68C/A93C/C97A/T115C, pH4 | | Descriptor: | Endolysin, GLYCEROL, HEXANE-1,6-DIOL, ... | | Authors: | Tamada, T, Hiromoto, T. | | Deposit date: | 2022-03-30 | | Release date: | 2023-03-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Creation of Cross-Linked Crystals With Intermolecular Disulfide Bonds Connecting Symmetry-Related Molecules Allows Retention of Tertiary Structure in Different Solvent Conditions.
Front Mol Biosci, 9, 2022
|
|
3HGN
 
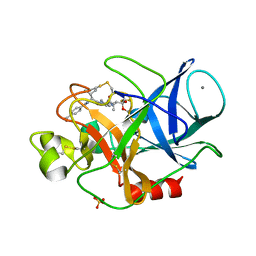 | | Structure of porcine pancreatic elastase complexed with a potent peptidyl inhibitor FR130180 determined by neutron crystallography | | Descriptor: | 4-[[(2S)-3-methyl-1-oxo-1-[(2S)-2-[[(3S)-1,1,1-trifluoro-4-methyl-2-oxo-pentan-3-yl]carbamoyl]pyrrolidin-1-yl]butan-2-yl]carbamoyl]benzoic acid, CALCIUM ION, Elastase-1, ... | | Authors: | Tamada, T, Kinoshita, T, Kuroki, R, Tada, T. | | Deposit date: | 2009-05-14 | | Release date: | 2009-07-28 | | Last modified: | 2024-10-30 | | Method: | NEUTRON DIFFRACTION (1.65 Å), X-RAY DIFFRACTION | | Cite: | Combined High-Resolution Neutron and X-ray Analysis of Inhibited Elastase Confirms the Active-Site Oxyanion Hole but Rules against a Low-Barrier Hydrogen Bond
J.Am.Chem.Soc., 131, 2009
|
|
3HGP
 
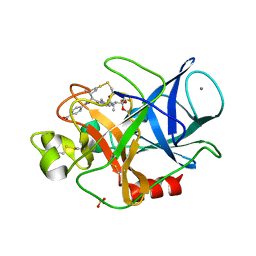 | | Structure of porcine pancreatic elastase complexed with a potent peptidyl inhibitor FR130180 determined by high resolution crystallography | | Descriptor: | 4-[[(2S)-3-methyl-1-oxo-1-[(2S)-2-[[(3S)-1,1,1-trifluoro-4-methyl-2-oxo-pentan-3-yl]carbamoyl]pyrrolidin-1-yl]butan-2-yl]carbamoyl]benzoic acid, CALCIUM ION, Elastase-1, ... | | Authors: | Tamada, T, Kinoshita, T, Kuroki, R, Tada, T. | | Deposit date: | 2009-05-14 | | Release date: | 2009-07-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (0.94 Å) | | Cite: | Combined High-Resolution Neutron and X-ray Analysis of Inhibited Elastase Confirms the Active-Site Oxyanion Hole but Rules against a Low-Barrier Hydrogen Bond
J.Am.Chem.Soc., 131, 2009
|
|
2D9Q
 
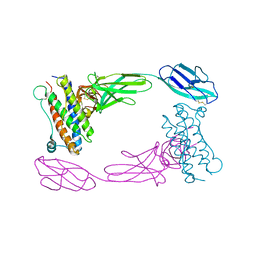 | | Crystal Structure of the Human GCSF-Receptor Signaling Complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CSF3, Granulocyte colony-stimulating factor receptor | | Authors: | Tamada, T, Kuroki, R. | | Deposit date: | 2005-12-12 | | Release date: | 2006-02-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Homodimeric cross-over structure of the human granulocyte colony-stimulating factor (GCSF) receptor signaling complex
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
7CQY
 
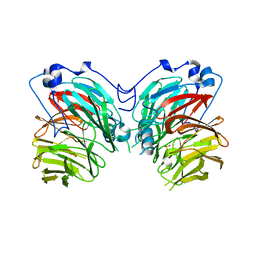 | |
1IUQ
 
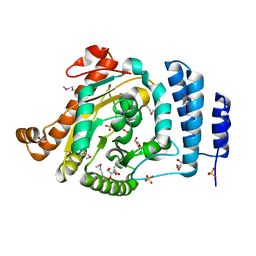 | | The 1.55 A Crystal Structure of Glycerol-3-Phosphate Acyltransferase | | Descriptor: | GLYCEROL, Glycerol-3-Phosphate Acyltransferase, SULFATE ION | | Authors: | Tamada, T, Feese, M.D, Kato, Y, Kuroki, R. | | Deposit date: | 2002-03-06 | | Release date: | 2003-10-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Substrate recognition and selectivity of plant glycerol-3-phosphate acyltransferases (GPATs) from Cucurbita moscata and Spinacea oleracea.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
6L8A
 
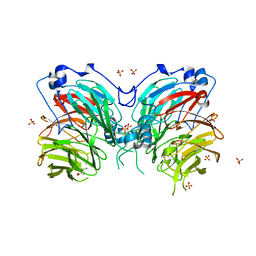 | | Tetrathionate hydrolase from Acidithiobacillus ferrooxidans | | Descriptor: | BETA-ALANINE, GLYCINE, SULFATE ION, ... | | Authors: | Tamada, T, Hirano, Y. | | Deposit date: | 2019-11-05 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.95004809 Å) | | Cite: | Reaction mechanism of tetrathionate hydrolysis based on the crystal structure of tetrathionate hydrolase from Acidithiobacillus ferrooxidans.
Protein Sci., 30, 2020
|
|
1IV9
 
 | |
1IV7
 
 | |
5ZCR
 
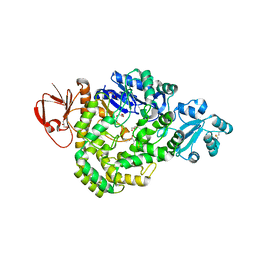 | | DSM5389 glycosyltrehalose synthase | | Descriptor: | GLYCEROL, MAGNESIUM ION, Maltooligosyl trehalose synthase | | Authors: | Tamada, T, Okazaki, N. | | Deposit date: | 2018-02-20 | | Release date: | 2018-11-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of glycosyltrehalose synthase from Sulfolobus shibatae DSM5389
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
2ZIK
 
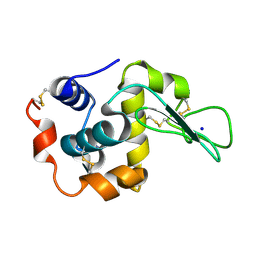 | |
2ZIJ
 
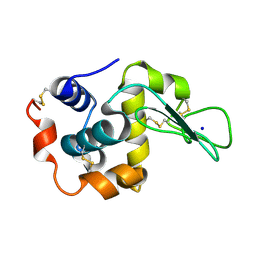 | |
2ZIL
 
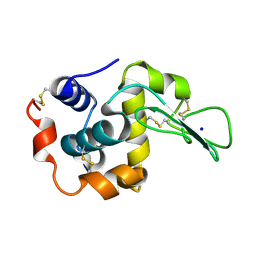 | | Crystal Structure of Human Lysozyme from Urine | | Descriptor: | Lysozyme C, SODIUM ION | | Authors: | Tamada, T, Kuroki, R, Koshiba, T. | | Deposit date: | 2008-02-18 | | Release date: | 2009-01-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Preparation and characterization of methyonyl-lysine attached human lysozyme expressed in Escherichia coli and its effective conversion to the authentic-like protein
To be Published
|
|
3X3G
 
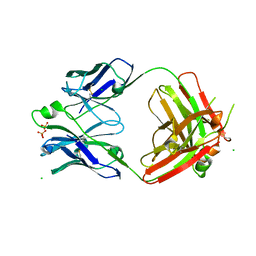 | | Fab fragment from anti TRAIL-R2 Human Agonist Antibody KMTR2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Heavy chain of KMTR2, ... | | Authors: | Tamada, T. | | Deposit date: | 2015-01-20 | | Release date: | 2015-12-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | TRAIL-R2 Superoligomerization Induced by Human Monoclonal Agonistic Antibody KMTR2
Sci Rep, 5, 2015
|
|
3X3F
 
 | |
8W6X
 
 | | Neutron structure of [NiFe]-hydrogenase from D. vulgaris Miyazaki F in its oxidized state | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, FE3-S4 CLUSTER, ... | | Authors: | Hiromoto, T, Tamada, T. | | Deposit date: | 2023-08-30 | | Release date: | 2023-09-13 | | Last modified: | 2023-11-15 | | Method: | NEUTRON DIFFRACTION (1.04 Å), X-RAY DIFFRACTION | | Cite: | New insights into the oxidation process from neutron and X-ray crystal structures of an O 2 -sensitive [NiFe]-hydrogenase.
Chem Sci, 14, 2023
|
|
7VOS
 
 | | High-resolution neutron and X-ray joint refined structure of high-potential iron-sulfur protein in the oxidized state | | Descriptor: | AMMONIUM ION, GLYCEROL, High-potential iron-sulfur protein, ... | | Authors: | Hanazono, Y, Hirano, Y, Takeda, K, Kusaka, K, Tamada, T, Miki, K. | | Deposit date: | 2021-10-14 | | Release date: | 2022-06-01 | | Last modified: | 2024-04-03 | | Method: | NEUTRON DIFFRACTION (0.66 Å), X-RAY DIFFRACTION | | Cite: | Revisiting the concept of peptide bond planarity in an iron-sulfur protein by neutron structure analysis.
Sci Adv, 8, 2022
|
|
4WHM
 
 | | Crystal structure of UDP-glucose: anthocyanidin 3-O-glucosyltransferase in complex with UDP | | Descriptor: | ACETATE ION, GLYCEROL, UDP-glucose:anthocyanidin 3-O-glucosyltransferase, ... | | Authors: | Hiromoto, T, Honjo, E, Tamada, T, Kuroki, R. | | Deposit date: | 2014-09-23 | | Release date: | 2015-01-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.851 Å) | | Cite: | Structural basis for acceptor-substrate recognition of UDP-glucose: anthocyanidin 3-O-glucosyltransferase from Clitoria ternatea
Protein Sci., 24, 2015
|
|
7XVX
 
 | |
8IHW
 
 | | X-ray crystal structure of D43R mutant of endo-1,4-beta glucanase from Eisenia fetida | | Descriptor: | CALCIUM ION, Endoglucanase, GLYCEROL, ... | | Authors: | Kuroki, C, Hirano, Y, Nakazawa, M, Sakamoto, T, Tamada, T, Ueda, M. | | Deposit date: | 2023-02-24 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A single mutation Asp43Arg was increased 2.5-fold the catalytic activity and maintained the stability of cold-adapted endo-1,4-beta glucanase (Ef-EG2) from Eisenia fetida.
Curr Res Biotechnol, 5, 2023
|
|
8IHX
 
 | | X-ray crystal structure of N372D mutant of endo-1,4-beta glucanase from Eisenia fetida | | Descriptor: | CALCIUM ION, Endoglucanase, GLYCEROL, ... | | Authors: | Kuroki, C, Hirano, Y, Nakazawa, M, Sakamoto, T, Tamada, T, Ueda, M. | | Deposit date: | 2023-02-24 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A single mutation Asp43Arg was increased 2.5-fold the catalytic activity and maintained the stability of cold-adapted endo-1,4-beta glucanase (Ef-EG2) from Eisenia fetida.
Curr Res Biotechnol, 5, 2023
|
|
