5DHD
 
 | | Crystal structure of ChBD2 from Thermococcus kodakarensis KOD1 | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, Chitinase, SULFATE ION | | Authors: | Hibi, M, Niwa, S, Takeda, K, Miki, K. | | Deposit date: | 2015-08-30 | | Release date: | 2016-02-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Crystal structures of chitin binding domains of chitinase from Thermococcus kodakarensis KOD1
Febs Lett., 590, 2016
|
|
3O0T
 
 | | Crystal structure of human phosphoglycerate mutase family member 5 (PGAM5) in complex with phosphate | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHATE ION, Serine/threonine-protein phosphatase PGAM5, ... | | Authors: | Chaikuad, A, Alfano, I, Picaud, S, Filippakopoulos, P, Barr, A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Takeda, K, Ichijo, H, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-07-20 | | Release date: | 2010-10-06 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of PGAM5 Provide Insight into Active Site Plasticity and Multimeric Assembly.
Structure, 25, 2017
|
|
5ZUI
 
 | | Crystal Structure of HSP104 from Chaetomium thermophilum | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Heat Shock Protein 104, SULFATE ION | | Authors: | Hanazono, Y, Inoue, Y, Noguchi, K, Yohda, M, Shinohara, K, Takeda, K, Miki, K. | | Deposit date: | 2018-05-07 | | Release date: | 2019-06-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Split conformation of Chaetomium thermophilum Hsp104 disaggregase.
Structure, 2021
|
|
3MXO
 
 | | Crystal structure oh human phosphoglycerate mutase family member 5 (PGAM5) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Chaikuad, A, Alfano, I, Picaud, S, Filippakopoulos, P, Barr, A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Takeda, K, Ichijo, H, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-05-07 | | Release date: | 2010-09-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of PGAM5 Provide Insight into Active Site Plasticity and Multimeric Assembly.
Structure, 25, 2017
|
|
5B3P
 
 | |
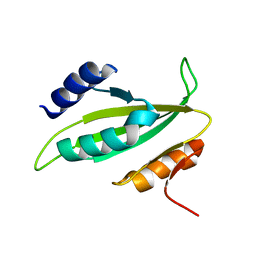
5B3Q
 
 | |
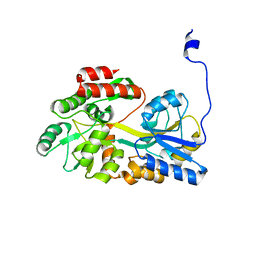
5B3Y
 
 | |
5B3W
 
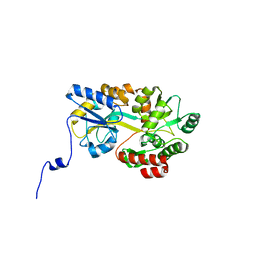 | | Crystal structure of hPin1 WW domain (5-15) fused with maltose-binding protein in C2221 form | | Descriptor: | CITRIC ACID, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1,Maltose-binding periplasmic protein, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Hanazono, Y, Takeda, K, Miki, K. | | Deposit date: | 2016-03-17 | | Release date: | 2016-10-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural studies of the N-terminal fragments of the WW domain: Insights into co-translational folding of a beta-sheet protein
Sci Rep, 6, 2016
|
|
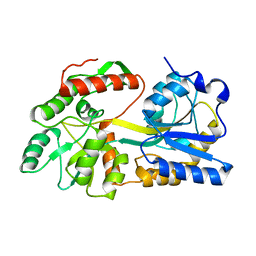
5B3X
 
 | |
5BMY
 
 | |
5B3Z
 
 | |
5WQR
 
 | | High resolution structure of high-potential iron-sulfur protein in the reduced state | | Descriptor: | GLYCEROL, High-potential iron-sulfur protein, IRON/SULFUR CLUSTER, ... | | Authors: | Ohno, H, Takeda, K, Niwa, S, Tsujinaka, T, Hanazono, Y, Hirano, Y, Miki, K. | | Deposit date: | 2016-11-28 | | Release date: | 2017-06-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (0.8 Å) | | Cite: | Crystallographic characterization of the high-potential iron-sulfur protein in the oxidized state at 0.8 angstrom resolution
PLoS ONE, 12, 2017
|
|
5WQQ
 
 | | High resolution structure of high-potential iron-sulfur protein in the oxidized state | | Descriptor: | GLYCEROL, High-potential iron-sulfur protein, IRON/SULFUR CLUSTER, ... | | Authors: | Ohno, H, Takeda, K, Niwa, S, Tsujinaka, T, Hanazono, Y, Hirano, Y, Miki, K. | | Deposit date: | 2016-11-28 | | Release date: | 2017-06-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (0.8 Å) | | Cite: | Crystallographic characterization of the high-potential iron-sulfur protein in the oxidized state at 0.8 angstrom resolution
PLoS ONE, 12, 2017
|
|
5ZIL
 
 | | Crystal structure of bacteriorhodopsin at 1.29 A resolution | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, RETINAL | | Authors: | Hasegawa, N, Jonotsuka, H, Miki, K, Takeda, K. | | Deposit date: | 2018-03-16 | | Release date: | 2018-10-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | X-ray structure analysis of bacteriorhodopsin at 1.3 angstrom resolution.
Sci Rep, 8, 2018
|
|
5ZIM
 
 | | Crystal structure of bacteriorhodopsin at 1.25 A resolution | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, RETINAL | | Authors: | Hasegawa, N, Jonotsuka, H, Miki, K, Takeda, K. | | Deposit date: | 2018-03-16 | | Release date: | 2018-10-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | X-ray structure analysis of bacteriorhodopsin at 1.3 angstrom resolution.
Sci Rep, 8, 2018
|
|
5ZIN
 
 | | Crystal structure of bacteriorhodopsin at 1.27 A resolution | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, RETINAL | | Authors: | Hasegawa, N, Jonotsuka, H, Miki, K, Takeda, K. | | Deposit date: | 2018-03-16 | | Release date: | 2018-10-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | X-ray structure analysis of bacteriorhodopsin at 1.3 angstrom resolution.
Sci Rep, 8, 2018
|
|
6AIR
 
 | | High resolution structure of perdeuterated high-potential iron-sulfur protein | | Descriptor: | GLYCEROL, High-potential iron-sulfur protein, IRON/SULFUR CLUSTER, ... | | Authors: | Hanazono, Y, Takeda, K, Miki, K. | | Deposit date: | 2018-08-24 | | Release date: | 2019-08-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | Characterization of perdeuterated high-potential iron-sulfur protein with high-resolution X-ray crystallography.
Proteins, 88, 2020
|
|
6AIQ
 
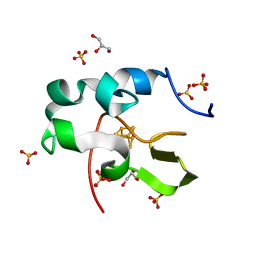 | | High resolution structure of recombinant high-potential iron-sulfur protein | | Descriptor: | GLYCEROL, High-potential iron-sulfur protein, IRON/SULFUR CLUSTER, ... | | Authors: | Hanazono, Y, Takeda, K, Miki, K. | | Deposit date: | 2018-08-24 | | Release date: | 2019-08-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | Characterization of perdeuterated high-potential iron-sulfur protein with high-resolution X-ray crystallography.
Proteins, 88, 2020
|
|
5ZCA
 
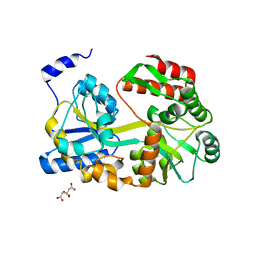 | | Crystal structure of lambda repressor (1-20) fused with maltose-binding protein | | Descriptor: | CITRIC ACID, Repressor protein cI,Maltose-binding periplasmic protein, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Hanazono, Y, Takeda, K, Miki, K. | | Deposit date: | 2018-02-16 | | Release date: | 2018-08-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Co-translational folding of alpha-helical proteins: structural studies of intermediate-length variants of the lambda repressor
Febs Open Bio, 8, 2018
|
|
6A2J
 
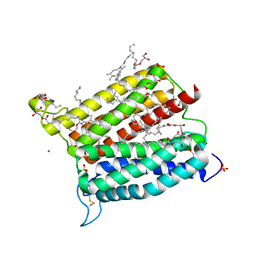 | | Crystal structure of heme A synthase from Bacillus subtilis | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, COPPER (II) ION, Heme A synthase, ... | | Authors: | Niwa, S, Takeda, K, Kosugi, M, Tsutsumi, E, Miki, K. | | Deposit date: | 2018-06-12 | | Release date: | 2018-11-21 | | Last modified: | 2018-12-12 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of heme A synthase fromBacillus subtilis.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6JGH
 
 | | Crystal structure of the F99S/M153T/V163A/T203I variant of GFP at 0.94 A | | Descriptor: | CHLORIDE ION, Green fluorescent protein | | Authors: | Eki, H, Tai, Y, Takaba, K, Hanazono, Y, Miki, K, Takeda, K. | | Deposit date: | 2019-02-14 | | Release date: | 2019-04-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (0.94 Å) | | Cite: | Subatomic resolution X-ray structures of green fluorescent protein.
Iucrj, 6, 2019
|
|
6JGI
 
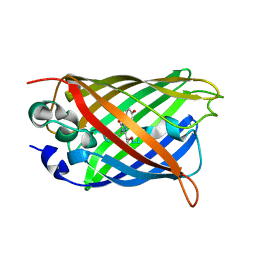 | | Crystal structure of the S65T/F99S/M153T/V163A variant of GFP at 0.85 A | | Descriptor: | Green fluorescent protein | | Authors: | Tai, Y, Takaba, K, Hanazono, Y, Miki, K, Takeda, K. | | Deposit date: | 2019-02-14 | | Release date: | 2019-04-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | Subatomic resolution X-ray structures of green fluorescent protein.
Iucrj, 6, 2019
|
|
6KKZ
 
 | | Crystal structure of the S65T/F99S/M153T/V163A variant of perdeuterated GFP at pD 8.5 | | Descriptor: | Green fluorescent protein | | Authors: | Tai, Y, Takaba, K, Hanazono, Y, Dao, H.A, Miki, K, Takeda, K. | | Deposit date: | 2019-07-28 | | Release date: | 2019-12-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | X-ray crystallographic studies on the hydrogen isotope effects of green fluorescent protein at sub-angstrom resolutions
Acta Crystallogr.,Sect.D, 75, 2019
|
|
6KL1
 
 | | Crystal structure of the S65T/F99S/M153T/V163A variant of non-deuterated GFP at pD 8.5 | | Descriptor: | Green fluorescent protein | | Authors: | Tai, Y, Takaba, K, Hanazono, Y, Dao, H.A, Miki, K, Takeda, K. | | Deposit date: | 2019-07-28 | | Release date: | 2019-12-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (0.851 Å) | | Cite: | X-ray crystallographic studies on the hydrogen isotope effects of green fluorescent protein at sub-angstrom resolutions
Acta Crystallogr.,Sect.D, 75, 2019
|
|
6KL0
 
 | | Crystal structure of the S65T/F99S/M153T/V163A variant of perdeuterated GFP at pD 7.0 | | Descriptor: | Green fluorescent protein | | Authors: | Tai, Y, Takaba, K, Hanazono, Y, Miki, K, Takeda, K. | | Deposit date: | 2019-07-28 | | Release date: | 2019-12-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (0.798 Å) | | Cite: | X-ray crystallographic studies on the hydrogen isotope effects of green fluorescent protein at sub-angstrom resolutions
Acta Crystallogr.,Sect.D, 75, 2019
|
|
