3O26
 
 | |
1TYK
 
 | | SOLUTION STRUCTURE OF A TOXIN FROM THE TARANTULA, GRAMMOSTOLA SPATULATA, WHICH INHIBITS MECHANOSENSITIVE ION CHANNELS | | Descriptor: | Toxin GsMTx-4 | | Authors: | Oswald, R.E, Suchyna, T.M, Mcfeeters, R, Gottlieb, P, Sachs, F. | | Deposit date: | 2004-07-08 | | Release date: | 2004-07-13 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Peptide Toxins that Block Mechanosensitive Ion Channels
J.Biol.Chem., 277, 2002
|
|
3O6W
 
 | | Crystal structure of monomeric KlHxk1 in crystal form VIII (open state) | | Descriptor: | GLYCEROL, Hexokinase, PHOSPHATE ION | | Authors: | Kuettner, E.B, Kettner, K, Keim, A, Kriegel, T.M, Strater, N. | | Deposit date: | 2010-07-29 | | Release date: | 2010-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystal Structure of Hexokinase KlHxk1 of Kluyveromyces lactis: A MOLECULAR BASIS FOR UNDERSTANDING THE CONTROL OF YEAST HEXOKINASE FUNCTIONS VIA COVALENT MODIFICATION AND OLIGOMERIZATION.
J.Biol.Chem., 285, 2010
|
|
3MSP
 
 | | MOTILE MAJOR SPERM PROTEIN (MSP) OF ASCARIS SUUM, NMR, 20 STRUCTURES | | Descriptor: | MAJOR SPERM PROTEIN | | Authors: | Haaf, A, Leclaire III, L, Roberts, G, Kent, H.M, Roberts, T.M, Stewart, M, Neuhaus, D. | | Deposit date: | 1998-09-10 | | Release date: | 1999-04-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the motile major sperm protein (MSP) of Ascaris suum - evidence for two manganese binding sites and the possible role of divalent cations in filament formation.
J.Mol.Biol., 284, 1998
|
|
3O4W
 
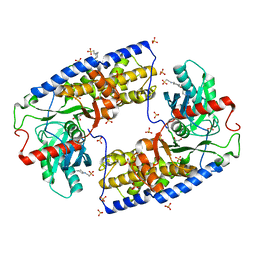 | | Crystal structure of dimeric KlHxk1 in crystal form IV | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, GLYCEROL, Hexokinase, ... | | Authors: | Kuettner, E.B, Kettner, K, Keim, A, Kriegel, T.M, Strater, N. | | Deposit date: | 2010-07-27 | | Release date: | 2010-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Crystal Structure of Hexokinase KlHxk1 of Kluyveromyces lactis: A MOLECULAR BASIS FOR UNDERSTANDING THE CONTROL OF YEAST HEXOKINASE FUNCTIONS VIA COVALENT MODIFICATION AND OLIGOMERIZATION.
J.Biol.Chem., 285, 2010
|
|
3O08
 
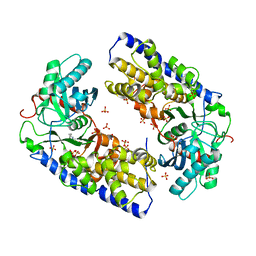 | | Crystal structure of dimeric KlHxk1 in crystal form I | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Hexokinase, SULFATE ION | | Authors: | Kuettner, E.B, Kettner, K, Keim, A, Kriegel, T.M, Strater, N. | | Deposit date: | 2010-07-19 | | Release date: | 2010-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Hexokinase KlHxk1 of Kluyveromyces lactis: A MOLECULAR BASIS FOR UNDERSTANDING THE CONTROL OF YEAST HEXOKINASE FUNCTIONS VIA COVALENT MODIFICATION AND OLIGOMERIZATION.
J.Biol.Chem., 285, 2010
|
|
3O8M
 
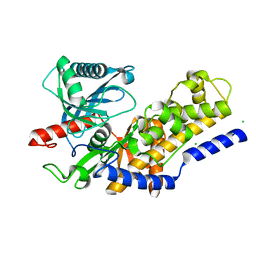 | | Crystal structure of monomeric KlHxk1 in crystal form XI with glucose bound (closed state) | | Descriptor: | CHLORIDE ION, Hexokinase, alpha-D-glucopyranose, ... | | Authors: | Kuettner, E.B, Kettner, K, Keim, A, Kriegel, T.M, Strater, N. | | Deposit date: | 2010-08-03 | | Release date: | 2010-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Crystal Structure of Hexokinase KlHxk1 of Kluyveromyces lactis: A MOLECULAR BASIS FOR UNDERSTANDING THE CONTROL OF YEAST HEXOKINASE FUNCTIONS VIA COVALENT MODIFICATION AND OLIGOMERIZATION.
J.Biol.Chem., 285, 2010
|
|
3O1W
 
 | | Crystal structure of dimeric KlHxk1 in crystal form III | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, GLYCEROL, Hexokinase, ... | | Authors: | Kuettner, E.B, Kettner, K, Keim, A, Kriegel, T.M, Strater, N. | | Deposit date: | 2010-07-22 | | Release date: | 2010-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Crystal Structure of Hexokinase KlHxk1 of Kluyveromyces lactis: A MOLECULAR BASIS FOR UNDERSTANDING THE CONTROL OF YEAST HEXOKINASE FUNCTIONS VIA COVALENT MODIFICATION AND OLIGOMERIZATION.
J.Biol.Chem., 285, 2010
|
|
3O80
 
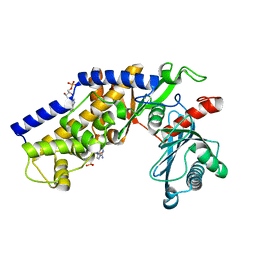 | | Crystal structure of monomeric KlHxk1 in crystal form IX (open state) | | Descriptor: | Hexokinase, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Kuettner, E.B, Kettner, K, Keim, A, Kriegel, T.M, Strater, N. | | Deposit date: | 2010-08-02 | | Release date: | 2010-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Crystal Structure of Hexokinase KlHxk1 of Kluyveromyces lactis: A MOLECULAR BASIS FOR UNDERSTANDING THE CONTROL OF YEAST HEXOKINASE FUNCTIONS VIA COVALENT MODIFICATION AND OLIGOMERIZATION.
J.Biol.Chem., 285, 2010
|
|
1UZ1
 
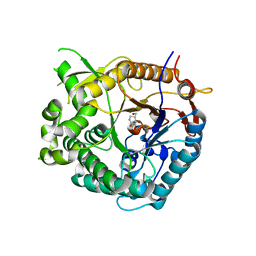 | | Family 1 b-glucosidase from Thermotoga maritima in complex with isofagomine lactam | | Descriptor: | (3S,4R,5R)-3,4-DIHYDROXY-5-(HYDROXYMETHYL)PIPERIDIN-2-ONE, BETA-GLUCOSIDASE A | | Authors: | Gloster, T.M, Macdonald, J, Stick, R.V, Davies, G.J. | | Deposit date: | 2004-03-03 | | Release date: | 2004-11-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Common Inhibition of Both -Glucosidases and -Mannosidases by Isofagomine Lactam Reflects Different Conformational Itineraries for Pyranoside Hydrolysis
Chembiochem, 5, 2004
|
|
1VQN
 
 | |
1VQ6
 
 | |
3LFG
 
 | |
1T1M
 
 | | Binding position of ribosome recycling factor (RRF) on the E. coli 70S ribosome | | Descriptor: | 42-mer fragment of double helix from 16S rRNA, dodecamer fragment of double helix from 23S rRNA, ribosome recycling factor | | Authors: | Agrawal, R.K, Sharma, M.R, Kiel, M.C, Hirokawa, G, Booth, T.M, Spahn, C.M, Grassucci, R.A, Kaji, A, Frank, J. | | Deposit date: | 2004-04-16 | | Release date: | 2004-06-15 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Visualization of ribosome-recycling factor on the Escherichia coli 70S ribosome: Functional implications
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
3LFJ
 
 | |
3LFU
 
 | | Crystal Structure of E. coli UvrD | | Descriptor: | DNA helicase II, SULFATE ION | | Authors: | Korolev, S, Waksman, G, Lohman, T.M. | | Deposit date: | 2010-01-18 | | Release date: | 2011-02-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Rotations of the 2B sub-domain of E. coli UvrD helicase/translocase coupled to nucleotide and DNA binding.
J.Mol.Biol., 411, 2011
|
|
1TFP
 
 | | TRANSTHYRETIN (FORMERLY KNOWN AS PREALBUMIN) | | Descriptor: | SULFATE ION, TRANSTHYRETIN | | Authors: | Sunde, M, Richardson, S.J, Chang, L, Pettersson, T.M, Schreiber, G, Blake, C.C.F. | | Deposit date: | 1996-01-05 | | Release date: | 1996-06-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The crystal structure of transthyretin from chicken.
Eur.J.Biochem., 236, 1996
|
|
1SRU
 
 | | Crystal structure of full length E. coli SSB protein | | Descriptor: | Single-strand binding protein | | Authors: | Savvides, S.N, Raghunathan, S, Fuetterer, K, Kozlov, A.G, Lohman, T.M, Waksman, G. | | Deposit date: | 2004-03-23 | | Release date: | 2004-08-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The C-terminal domain of full-length E. coli SSB is disordered even when bound to DNA.
Protein Sci., 13, 2004
|
|
1QYZ
 
 | | Characterization of the malformed, recombinant cytochrome rC552 | | Descriptor: | 2-ACETYL-PROTOPORPHYRIN IX, Cytochrome c-552 | | Authors: | Fee, J.A, Todaro, T.R, Luna, E, Sanders, D, Hunsicker-Wang, L.M, Patel, K.M, Bren, K.L, Gomez-Moran, E, Hill, M.G, Ai, J, Loehr, T.M, Oertling, W.A, Williams, P.A, Stout, C.D, McRee, D, Pastuszyn, A. | | Deposit date: | 2003-09-12 | | Release date: | 2004-09-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Cytochrome rC552, formed during expression of the truncated, Thermus thermophilus cytochrome c552 gene in the cytoplasm of Escherichia coli, reacts spontaneously to form protein-bound 2-formyl-4-vinyl (Spirographis) heme.
Biochemistry, 43, 2004
|
|
3L7R
 
 | | crystal structure of MetE from streptococcus mutans | | Descriptor: | 5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase, SULFATE ION, ZINC ION | | Authors: | Fu, T.M, Liang, Y.H, Su, X.D. | | Deposit date: | 2009-12-29 | | Release date: | 2011-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.397 Å) | | Cite: | Crystal Structures of Cobalamin-Independent Methionine Synthase (MetE) from Streptococcus mutans: A Dynamic Zinc-Inversion Model
J.Mol.Biol., 412, 2011
|
|
3L7W
 
 | |
3LFH
 
 | |
3L8A
 
 | |
1UD7
 
 | | SOLUTION STRUCTURE OF THE DESIGNED HYDROPHOBIC CORE MUTANT OF UBIQUITIN, 1D7 | | Descriptor: | PROTEIN (UBIQUITIN CORE MUTANT 1D7) | | Authors: | Johnson, E.C, Lazar, G.A, Desjarlais, J.R, Handel, T.M. | | Deposit date: | 1999-04-07 | | Release date: | 1999-05-06 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of a designed hydrophobic core variant of ubiquitin.
Structure Fold.Des., 7, 1999
|
|
1S18
 
 | | Structure and protein design of human apyrase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Dai, J, Liu, J, Deng, Y, Smith, T.M, Lu, M. | | Deposit date: | 2004-01-05 | | Release date: | 2004-03-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and protein design of a human platelet function inhibitor.
Cell(Cambridge,Mass.), 116, 2004
|
|
