2AB2
 
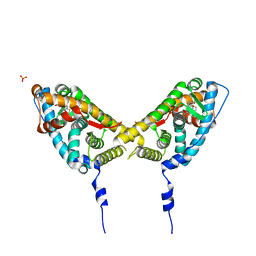 | | Mineralocorticoid Receptor Double Mutant with Bound Spironolactone | | Descriptor: | Mineralocorticoid receptor, SPIRONOLACTONE, SULFATE ION | | Authors: | Bledsoe, R.K, Madauss, K.P, Holt, J.A, Apolito, C.J, Lambert, M.H, Pearce, K.H, Stanley, T.B, Stewart, E.L, Trump, R.P, Willson, T.M, Williams, S.P. | | Deposit date: | 2005-07-14 | | Release date: | 2005-07-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A Ligand-mediated Hydrogen Bond Network Required for the Activation of the Mineralocorticoid Receptor
J.Biol.Chem., 280, 2005
|
|
2HLI
 
 | | Solution structure of Crotonaldehyde-Derived N2-[3-Oxo-1(S)-methyl-propyl]-dG DNA Adduct in the 5'-CpG-3' Sequence | | Descriptor: | DNA dodecamer, DNA dodecamer with S-crotonaldehyde adduct | | Authors: | Cho, Y.-J, Wang, H, Kozekov, I.D, Kurtz, A.J, Jacob, J, Voehler, M, Smith, J, Harris, T.M, Rizzo, C.J, Stone, M.P. | | Deposit date: | 2006-07-07 | | Release date: | 2006-09-19 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Orientation of the Crotonaldehyde-Derived N(2)-[3-Oxo-1(S)-methyl-propyl]-dG DNA Adduct Hinders Interstrand Cross-Link Formation in the 5'-CpG-3' Sequence
Chem.Res.Toxicol., 19, 2006
|
|
2A13
 
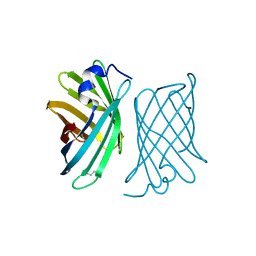 | | X-ray structure of protein from Arabidopsis thaliana AT1G79260 | | Descriptor: | At1g79260 | | Authors: | Wesenberg, G.E, Phillips Jr, G.N, McCoy, J.G, Bitto, E, Bingman, C.A, Allard, S.T.M, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2005-06-17 | | Release date: | 2005-07-12 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | The structure and NO binding properties of the nitrophorin-like heme-binding protein from Arabidopsis thaliana gene locus At1g79260.1.
Proteins, 78, 2010
|
|
2HMD
 
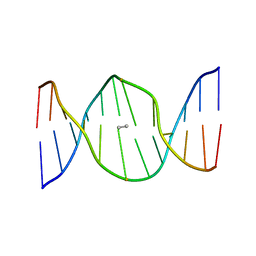 | | Stereochemistry Modulates Stability of Reduced Inter-Strand Cross-Links Arising From R- and S-alpha-methyl-gamma-OH-1,N2-propano-2'-Deoxyguanine in the 5'-CpG-3' DNA Sequence | | Descriptor: | DNA dodecamer with interstrand cross-link | | Authors: | Cho, Y.-J, Kozekov, I.D, Harris, T.M, Rizzo, C.J, Stone, M.P. | | Deposit date: | 2006-07-11 | | Release date: | 2007-03-13 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Stereochemistry Modulates the Stability of Reduced Interstrand Cross-Links Arising from R- and S-alpha-CH(3)-gamma-OH-1,N(2)-Propano-2'-deoxyguanosine in the 5'-CpG-3' DNA Sequence
Biochemistry, 46, 2007
|
|
2A62
 
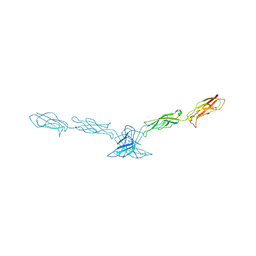 | | Crystal structure of mouse cadherin-8 EC1-3 | | Descriptor: | CALCIUM ION, Cadherin-8 | | Authors: | Patel, S.D, Ciatto, C, Chen, C.P, Bahna, F, Arkus, N, Schieren, I, Jessell, T.M, Honig, B, Price, S.R, Shapiro, L. | | Deposit date: | 2005-07-01 | | Release date: | 2006-04-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Type II cadherin ectodomain structures: implications for classical cadherin specificity.
Cell(Cambridge,Mass.), 124, 2006
|
|
2JG0
 
 | | Family 37 trehalase from Escherichia coli in complex with 1- thiatrehazolin | | Descriptor: | N-[(3aS,4R,5S,6S,6aS)-4,5,6-trihydroxy-4-(hydroxymethyl)-4,5,6,6a-tetrahydro-3aH-cyclopenta[d][1,3]thiazol-2-yl]-alpha- D-glucopyranosylamine, PERIPLASMIC TREHALASE | | Authors: | Gibson, R.P, Gloster, T.M, Roberts, S, Warren, R.A.J, Storch De Gracia, I, Garcia, A, Chiara, J.L, Davies, G.J. | | Deposit date: | 2007-02-07 | | Release date: | 2007-02-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Molecular Basis for Trehalase Inhibition Revealed by the Structure of Trehalase in Complex with Potent Inhibitors.
Angew.Chem.Int.Ed.Engl., 46, 2007
|
|
2JJB
 
 | | Family 37 trehalase from Escherichia coli in complex with casuarine-6- O-alpha-glucopyranose | | Descriptor: | 1,2-ETHANEDIOL, CASUARINE, PERIPLASMIC TREHALASE, ... | | Authors: | Gloster, T.M, Roberts, S, Davies, G.J, Cardona, F, Parmeggiani, C, Bonaccini, C, Gratteri, P, Sim, L, Rose, D.R, Goti, A. | | Deposit date: | 2008-03-28 | | Release date: | 2009-01-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Total Syntheses of Casuarine and its 6-O-Alpha-Glucoside: Complementary Inhibition Towards Glycoside Hydrolases of the Gh31 and Gh37 Families.
Chemistry, 15, 2009
|
|
1VKP
 
 | | X-RAY STRUCTURE OF GENE PRODUCT FROM ARABIDOPSIS THALIANA AT5G08170, AGMATINE IMINOHYDROLASE | | Descriptor: | 1,2-ETHANEDIOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, AGMATINE IMINOHYDROLASE, ... | | Authors: | Wesenberg, G.E, Smith, D.W, Phillips Jr, G.N, Bingman, C.A, Allard, S.T.M, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2004-06-15 | | Release date: | 2004-08-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | X-ray Structure of Gene Product from Arabidopsis Thaliana At5g08170
To be published
|
|
2JH1
 
 | | Crystal structure of Toxoplasma gondii micronemal protein 1 | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Blumenschein, T.M.A, Friedrich, N, Childs, R.A, Saouros, S, Carpenter, E.P, Campanero-Rhodes, M.A, Simpson, P, Koutroukides, T, Blackman, M.J, Feizi, T, Soldati-Favre, D, Matthews, S. | | Deposit date: | 2007-02-19 | | Release date: | 2007-05-22 | | Last modified: | 2019-01-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Atomic Resolution Insight Into Host Cell Recognition by Toxoplasma Gondii.
Embo J., 26, 2007
|
|
1VK5
 
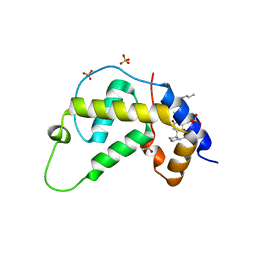 | | X-ray Structure of Gene Product from Arabidopsis Thaliana At3g22680 | | Descriptor: | 1,2-ETHANEDIOL, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, SULFATE ION, ... | | Authors: | Wesenberg, G.E, Smith, D.W, Phillips Jr, G.N, Johnson, K.A, Bingman, C.A, Allard, S.T.M, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2004-05-06 | | Release date: | 2004-05-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.604 Å) | | Cite: | Structure at 1.6 A resolution of the protein from gene locus At3g22680 from Arabidopsis thaliana.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
2LKV
 
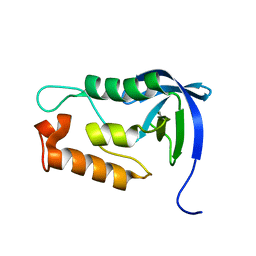 | | Staphylococcal Nuclease PHS variant | | Descriptor: | Thermonuclease | | Authors: | Matzapetakis, M, Pais, T.M, Lamosa, P, Turner, D.L, Santos, H. | | Deposit date: | 2011-10-21 | | Release date: | 2012-09-12 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Mannosylglycerate stabilizes staphylococcal nuclease with restriction of slow beta-sheet motions.
Protein Sci., 21, 2012
|
|
7RZZ
 
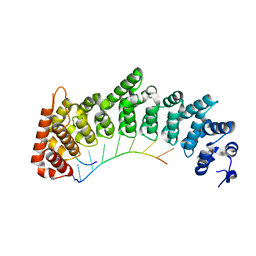 | |
7RR9
 
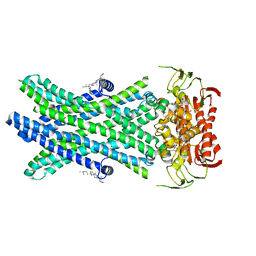 | | Cryo-EM Structure of Nanodisc reconstituted ABCD1 in nucleotide bound outward open conformation | | Descriptor: | ATP-binding cassette sub-family D member 1, CHOLESTEROL, MAGNESIUM ION, ... | | Authors: | Alam, A, Le, L.T.M, Thompson, J.R. | | Deposit date: | 2021-08-09 | | Release date: | 2022-01-19 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structures of the human peroxisomal fatty acid transporter ABCD1 in a lipid environment
Commun Biol, 5, 2022
|
|
7RRA
 
 | |
7S02
 
 | |
7TU0
 
 | | Structure of the L. blandensis dGTPase bound to Mn | | Descriptor: | MANGANESE (II) ION, SULFATE ION, dGTP triphosphohydrolase | | Authors: | Sikkema, A.P, Klemm, B.P, Horng, J.C, Hall, T.M.T. | | Deposit date: | 2022-02-02 | | Release date: | 2022-06-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | High-resolution structures of the SAMHD1 dGTPase homolog from Leeuwenhoekiella blandensis reveal a novel mechanism of allosteric activation by dATP.
J.Biol.Chem., 298, 2022
|
|
7TU1
 
 | | Structure of the L. blandensis dGTPase R37A mutant | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, dGTP triphosphohydrolase | | Authors: | Sikkema, A.P, Klemm, B.P, Horng, J.C, Hall, T.M.T. | | Deposit date: | 2022-02-02 | | Release date: | 2022-06-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High-resolution structures of the SAMHD1 dGTPase homolog from Leeuwenhoekiella blandensis reveal a novel mechanism of allosteric activation by dATP.
J.Biol.Chem., 298, 2022
|
|
7TU2
 
 | | Structure of the L. blandensis dGTPase R37A mutant bound to Mn | | Descriptor: | MANGANESE (II) ION, SULFATE ION, dGTP triphosphohydrolase | | Authors: | Sikkema, A.P, Klemm, B.P, Horng, J.C, Hall, T.M.T. | | Deposit date: | 2022-02-02 | | Release date: | 2022-06-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | High-resolution structures of the SAMHD1 dGTPase homolog from Leeuwenhoekiella blandensis reveal a novel mechanism of allosteric activation by dATP.
J.Biol.Chem., 298, 2022
|
|
7TU3
 
 | | Structure of the L. blandensis dGTPase del55-58 mutant | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Sikkema, A.P, Klemm, B.P, Horng, J.C, Hall, T.M.T. | | Deposit date: | 2022-02-02 | | Release date: | 2022-06-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | High-resolution structures of the SAMHD1 dGTPase homolog from Leeuwenhoekiella blandensis reveal a novel mechanism of allosteric activation by dATP.
J.Biol.Chem., 298, 2022
|
|
7TU4
 
 | | Structure of the L. blandensis dGTPase del55-58 mutant bound to Mn | | Descriptor: | 1,2-ETHANEDIOL, MANGANESE (II) ION, SULFATE ION, ... | | Authors: | Sikkema, A.P, Klemm, B.P, Horng, J.C, Hall, T.M.T. | | Deposit date: | 2022-02-02 | | Release date: | 2022-06-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | High-resolution structures of the SAMHD1 dGTPase homolog from Leeuwenhoekiella blandensis reveal a novel mechanism of allosteric activation by dATP.
J.Biol.Chem., 298, 2022
|
|
