1R2U
 
 | | NMR structure of the N domain of trout cardiac troponin C at 30 C | | 分子名称: | CALCIUM ION, troponin C | | 著者 | Blumenschein, T.M, Gillis, T.E, Tibbits, G.F, Sykes, B.D. | | 登録日 | 2003-09-29 | | 公開日 | 2004-06-08 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Effect of temperature on the structure of trout troponin C
Biochemistry, 43, 2004
|
|
1R6P
 
 | | NMR structure of the N-terminal domain of trout cardiac troponin C at 7 C | | 分子名称: | CALCIUM ION, troponin C | | 著者 | Blumenschein, T.M, Gillis, T.E, Tibbits, G.F, Sykes, B.D. | | 登録日 | 2003-10-15 | | 公開日 | 2004-06-08 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Effect of temperature on the structure of trout troponin C
Biochemistry, 43, 2004
|
|
1UNJ
 
 | | Crystal structure of a 7-Aminoactinomycin D complex with non-complementary DNA | | 分子名称: | 5'-D(*TP*TP*AP*GP*BRU*TP)-3', 7-AMINO-ACTINOMYCIN D | | 著者 | Alexopoulos, E.C, Klement, R, Jares-Erijman, E.A, Uson, I, Jovin, T.M, Sheldrick, G.M. | | 登録日 | 2003-09-10 | | 公開日 | 2004-12-16 | | 最終更新日 | 2024-07-10 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal and Solution Structures of 7-Amino-Actinomycin D Complexes with D(Ttagbrut), D(Ttagtt) and D(Tttagttt)
Acta Crystallogr.,Sect.D, 61, 2005
|
|
3NZ1
 
 | | Crystal Structure of Kemp Elimination Catalyst 1A53-2 Complexed with Transition State Analog 5-Nitro Benzotriazole | | 分子名称: | 5-nitro-1H-benzotriazole, Indole-3-glycerol phosphate synthase, L(+)-TARTARIC ACID, ... | | 著者 | Lee, T.M, Privett, H.K, Kaiser, J.T, Mayo, S.L. | | 登録日 | 2010-07-15 | | 公開日 | 2011-06-29 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.56 Å) | | 主引用文献 | Iterative approach to computational enzyme design.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1US3
 
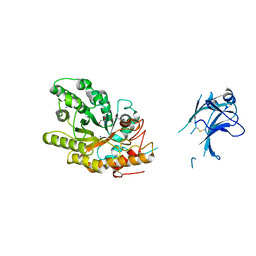 | | Native xylanase10C from Cellvibrio japonicus | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ENDO-BETA-1,4-XYLANASE PRECURSOR, GLYCEROL, ... | | 著者 | Pell, G, Szabo, L, Charnock, S.J, Xie, H, Gloster, T.M, Davies, G.J, Gilbert, H.J. | | 登録日 | 2003-11-17 | | 公開日 | 2003-12-18 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structural and Biochemical Analysis of Cellvibrio Japonicus Xylanase 10C: How Variation in Substrate-Binding Cleft Influences the Catalytic Profile of Family Gh-10 Xylanases
J.Biol.Chem., 279, 2004
|
|
1UQZ
 
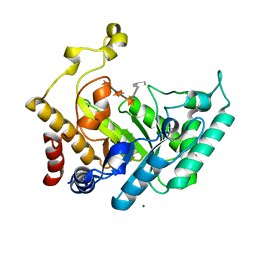 | | Xylanase Xyn10B mutant (E262S) from Cellvibrio mixtus in complex with 4-O-methyl glucuronic acid | | 分子名称: | 4-O-methyl-alpha-D-glucopyranuronic acid-(1-2)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, CHLORIDE ION, ENDOXYLANASE, ... | | 著者 | Pell, G, Taylor, E.J, Gloster, T.M, Turkenburg, J.P, Fontes, C.M.G.A, Ferreira, L.M.A, Davies, G.J, Gilbert, H.J. | | 登録日 | 2003-10-24 | | 公開日 | 2003-12-18 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | The Mechanisms by which Family 10 Glycoside Hydrolases Bind Decorated Substrates
J.Biol.Chem., 279, 2004
|
|
1V0M
 
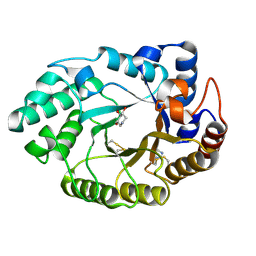 | | Xylanase Xyn10a from Streptomyces lividans in complex with xylobio-deoxynojirimycin at pH 7.5 | | 分子名称: | ENDO-1,4-BETA-XYLANASE A, IMIDAZOLE, PIPERIDINE-3,4,5-TRIOL, ... | | 著者 | Gloster, T.M, Williams, S.J, Roberts, S, Tarling, C.A, Wicki, J, Withers, S.G, Davies, G.J. | | 登録日 | 2004-03-31 | | 公開日 | 2004-08-16 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.07 Å) | | 主引用文献 | Atomic Resolution Analyses of the Binding of Xylobiose-Derived Deoxynojirimycin and Isofagomine to Xylanase Xyn10A
Chem.Commun.(Camb.), 16, 2004
|
|
1V0K
 
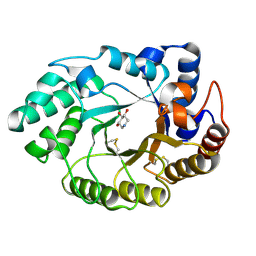 | | Xylanase Xyn10A from Streptomyces lividans in complex with xylobio-deoxynojirimycin at pH 5.8 | | 分子名称: | ENDO-1,4-BETA-XYLANASE A, PIPERIDINE-3,4,5-TRIOL, beta-D-xylopyranose | | 著者 | Gloster, T.M, Williams, S.J, Roberts, S, Tarling, C.A, Wicki S, J, Withers, G, Davies, G.J. | | 登録日 | 2004-03-31 | | 公開日 | 2004-08-16 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.03 Å) | | 主引用文献 | Atomic Resolution Analyses of the Binding of Xylobiose-Derived Deoxynojirimycin and Isofagomine to Xylanase Xyn10A
Chem.Commun.(Camb.), 16, 2004
|
|
3MXL
 
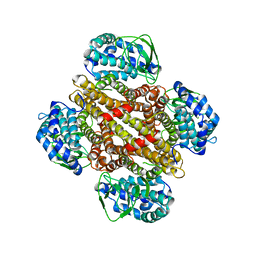 | |
1RAL
 
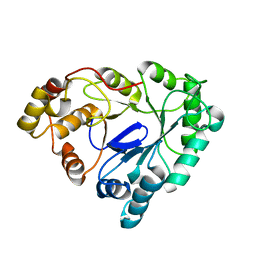 | | THREE-DIMENSIONAL STRUCTURE OF RAT LIVER 3ALPHA-HYDROXYSTEROID(SLASH)DIHYDRODIOL DEHYDROGENASE: A MEMBER OF THE ALDO-KETO REDUCTASE SUPERFAMILY | | 分子名称: | 3-ALPHA-HYDROXYSTEROID DEHYDROGENASE | | 著者 | Hoog, S.S, Pawlowski, J.E, Alzari, P.M, Penning, T.M, Lewis, M. | | 登録日 | 1994-02-04 | | 公開日 | 1994-04-30 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Three-dimensional structure of rat liver 3 alpha-hydroxysteroid/dihydrodiol dehydrogenase: a member of the aldo-keto reductase superfamily.
Proc.Natl.Acad.Sci.USA, 91, 1994
|
|
1UWT
 
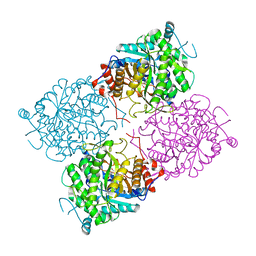 | | Structure of beta-glycosidase from Sulfolobus solfataricus in complex with D-galactohydroximo-1,5-lactam | | 分子名称: | (2E,3R,4R,5R,6S)-3,4,5-TRIHYDROXY-6-(HYDROXYMETHYL)-2-PIPERIDINONE, ACETATE ION, BETA-GALACTOSIDASE | | 著者 | Gloster, T.M, Roberts, S, Ducros, V.M.-A, Perugino, G, Rossi, M, Hoos, R, Moracci, M, Vasella, A, Davies, G.J. | | 登録日 | 2004-02-11 | | 公開日 | 2004-05-20 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structural studies of the beta-glycosidase from Sulfolobus solfataricus in complex with covalently and noncovalently bound inhibitors.
Biochemistry, 43, 2004
|
|
3OT9
 
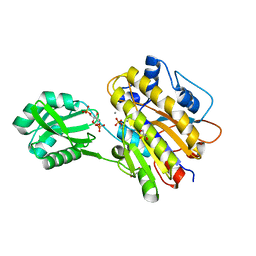 | | Phosphopentomutase from Bacillus cereus bound to glucose-1,6-bisphosphate | | 分子名称: | 1,6-di-O-phosphono-alpha-D-glucopyranose, GLYCEROL, MANGANESE (II) ION, ... | | 著者 | Panosian, T.D, Nannemann, D.P, Watkins, G, Phalen, V, Wadzinski, B, Bachmann, B.O, Iverson, T.M. | | 登録日 | 2010-09-10 | | 公開日 | 2010-12-29 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Bacillus cereus Phosphopentomutase Is an Alkaline Phosphatase Family Member That Exhibits an Altered Entry Point into the Catalytic Cycle.
J.Biol.Chem., 286, 2011
|
|
1UR2
 
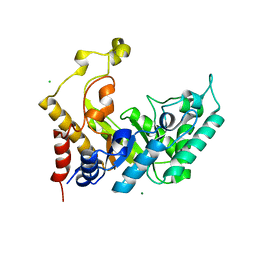 | | Xylanase Xyn10B mutant (E262S) from Cellvibrio mixtus in complex with arabinofuranose alpha 1,3 linked to xylotriose | | 分子名称: | CHLORIDE ION, ENDOXYLANASE, MAGNESIUM ION, ... | | 著者 | Pell, G, Taylor, E.J, Gloster, T.M, Turkenburg, J.P, Fontes, C.M.G.A, Ferreira, L.M.A, Davies, G.J, Gilbert, H.J. | | 登録日 | 2003-10-24 | | 公開日 | 2003-12-18 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | The Mechanisms by which Family 10 Glycoside Hydrolases Bind Decorated Substrates
J.Biol.Chem., 279, 2004
|
|
1UWS
 
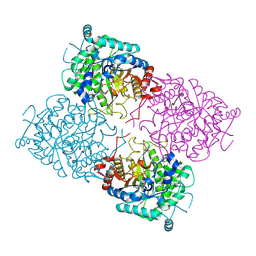 | | Structure of beta-glycosidase from Sulfolobus solfataricus in complex with 2-deoxy-2-fluoro-glucose | | 分子名称: | 2-deoxy-2-fluoro-alpha-D-glucopyranose, ACETATE ION, BETA-GALACTOSIDASE | | 著者 | Gloster, T.M, Roberts, S, Ducros, V.M.-A, Perugino, G, Rossi, M, Hoos, R, Moracci, M, Vasella, A, Davies, G.J. | | 登録日 | 2004-02-11 | | 公開日 | 2004-05-20 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structural studies of the beta-glycosidase from Sulfolobus solfataricus in complex with covalently and noncovalently bound inhibitors.
Biochemistry, 43, 2004
|
|
1V0N
 
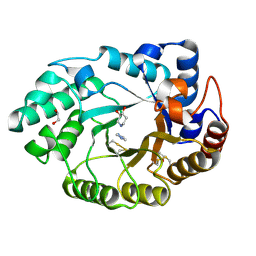 | | Xylanase Xyn10a from Streptomyces lividans in complex with xylobio-isofagomine at pH 7.5 | | 分子名称: | 1,2-ETHANEDIOL, ENDO-1,4-BETA-XYLANASE A, IMIDAZOLE, ... | | 著者 | Gloster, T.M, Williams, S.J, Roberts, S, Tarling, C.A, Wicki, J, Withers, S.G, Davies, G.J. | | 登録日 | 2004-03-31 | | 公開日 | 2004-08-16 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | Atomic Resolution Analyses of the Binding of Xylobiose-Derived Deoxynojirimycin and Isofagomine to Xylanase Xyn10A
Chem.Commun.(Camb.), 16, 2004
|
|
1UNM
 
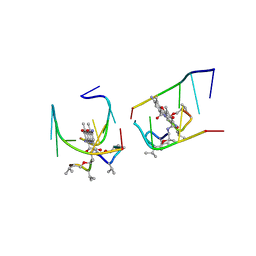 | | Crystal structure of 7-Aminoactinomycin D with non-complementary DNA | | 分子名称: | 5'-D(*TP*TP*AP*GP*BRU*TP)-3', 7-AMINOACTINOMYCIN D | | 著者 | Alexopoulos, E.C, Klement, R, Jares-Erijman, E.A, Uson, I, Jovin, T.M, Sheldrick, G.M. | | 登録日 | 2003-09-11 | | 公開日 | 2004-09-24 | | 最終更新日 | 2024-07-10 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal and Solution Structures of 7-Amino-Actinomycin D Complexes with D(Ttagbrut), D(Ttagtt) and D(Tttagttt)
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1URK
 
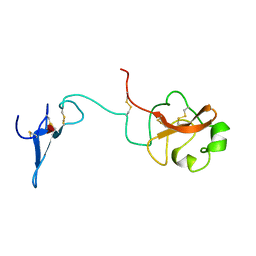 | | SOLUTION STRUCTURE OF THE AMINO TERMINAL FRAGMENT OF UROKINASE-TYPE PLASMINOGEN ACTIVATOR | | 分子名称: | PLASMINOGEN ACTIVATOR, alpha-L-fucopyranose | | 著者 | Hansen, A.P, Petros, A.M, Meadows, R.P, Nettesheim, D.G, Mazar, A.P, Olejniczak, E.T, Xu, R.X, Pederson, T.M, Henkin, J, Fesik, S.W. | | 登録日 | 1994-01-10 | | 公開日 | 1995-05-08 | | 最終更新日 | 2020-07-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the amino-terminal fragment of urokinase-type plasminogen activator.
Biochemistry, 33, 1994
|
|
1V0L
 
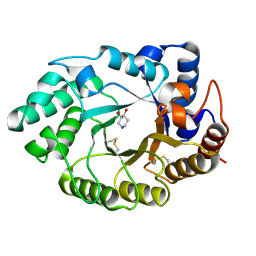 | | Xylanase Xyn10A from Streptomyces lividans in complex with xylobio-isofagomine at pH 5.8 | | 分子名称: | ENDO-1,4-BETA-XYLANASE A, PIPERIDINE-3,4-DIOL, beta-D-xylopyranose | | 著者 | Gloster, T.M, Williams, S.J, Roberts, S, Tarling, C.A, Wicki, J, Withers, S.G, Davies, G.J. | | 登録日 | 2004-03-31 | | 公開日 | 2004-08-16 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (0.98 Å) | | 主引用文献 | Atomic Resolution Analyses of the Binding of Xylobiose-Derived Deoxynojirimycin and Isofagomine to Xylanase Xyn10A
Chem.Commun.(Camb.), 16, 2004
|
|
1UR1
 
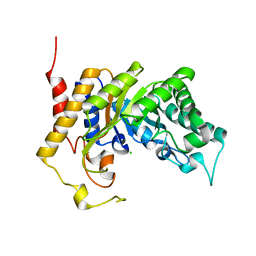 | | Xylanase Xyn10B mutant (E262S) from Cellvibrio mixtus in complex with arabinofuranose alpha-1,3 linked to xylobiose | | 分子名称: | CHLORIDE ION, ENDOXYLANASE, MAGNESIUM ION, ... | | 著者 | Pell, G, Taylor, E.J, Gloster, T.M, Turkenburg, J.P, Fontes, C.M.G.A, Ferreira, L.M.A, Davies, G.J, Gilbert, H.J. | | 登録日 | 2003-10-24 | | 公開日 | 2003-12-18 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.43 Å) | | 主引用文献 | The Mechanisms by which Family 10 Glycoside Hydrolases Bind Decorated Substrates
J.Biol.Chem., 279, 2004
|
|
1UQY
 
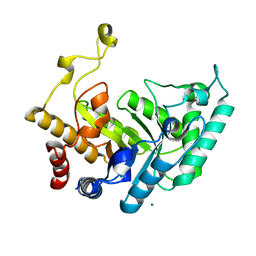 | | Xylanase Xyn10B mutant (E262S) from Cellvibrio mixtus in complex with xylopentaose | | 分子名称: | ENDOXYLANASE, MAGNESIUM ION, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-alpha-D-xylopyranose, ... | | 著者 | Pell, G, Taylor, E.J, Gloster, T.M, Turkenburg, J.P, Fontes, C.M.G.A, Ferreira, L.M.A, Davies, G.J, Gilbert, H.J. | | 登録日 | 2003-10-23 | | 公開日 | 2003-12-18 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.72 Å) | | 主引用文献 | The Mechanisms by which Family 10 Glycoside Hydrolases Bind Decorated Substrates
J.Biol.Chem., 279, 2004
|
|
1SPW
 
 | | Solution Structure of a Loop Truncated Mutant from D. gigas Rubredoxin, NMR | | 分子名称: | Rubredoxin | | 著者 | Pais, T.M, Lamosa, P, dos Santos, W, LeGall, J, Turner, D.L, Santos, H. | | 登録日 | 2004-03-17 | | 公開日 | 2005-03-29 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural determinants of protein stabilization by solutes: the importance of the hairpin loop in rubredoxins
FEBS J., 272, 2005
|
|
3PAR
 
 | | Surfactant Protein-A neck and carbohydrate recognition domain (NCRD) in the absence of ligand | | 分子名称: | CALCIUM ION, Pulmonary surfactant-associated protein A, SULFATE ION | | 著者 | Shang, F, Rynkiewicz, M.J, McCormack, F.X, Wu, H, Cafarella, T.M, Head, J, Seaton, B.A. | | 登録日 | 2010-10-19 | | 公開日 | 2010-11-03 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystallographic complexes of surfactant protein A and carbohydrates reveal ligand-induced conformational change.
J.Biol.Chem., 286, 2011
|
|
1US2
 
 | | Xylanase10C (mutant E385A) from Cellvibrio japonicus in complex with xylopentaose | | 分子名称: | ENDO-BETA-1,4-XYLANASE, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | 著者 | Pell, G, Szabo, L, Charnock, S.J, Xie, H, Gloster, T.M, Davies, G.J, Gilbert, H.J. | | 登録日 | 2003-11-17 | | 公開日 | 2003-12-18 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structural and Biochemical Analysis of Cellvibrio Japonicus Xylanase 10C: How Variation in Substrate-Binding Cleft Influences the Catalytic Profile of Family Gh-10 Xylanases
J.Biol.Chem., 279, 2004
|
|
3PAK
 
 | | Crystal Structure of Rat Surfactant Protein A neck and carbohydrate recognition domain (NCRD) complexed with Mannose | | 分子名称: | CALCIUM ION, Pulmonary surfactant-associated protein A, SODIUM ION, ... | | 著者 | Shang, F, Rynkiewicz, M.J, McCormack, F.X, Wu, H, Cafarella, T.M, Head, J, Seaton, B.A. | | 登録日 | 2010-10-19 | | 公開日 | 2010-11-03 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystallographic complexes of surfactant protein A and carbohydrates reveal ligand-induced conformational change.
J.Biol.Chem., 286, 2011
|
|
1T1O
 
 | | Components of the control 70S ribosome to provide reference for the RRF binding site | | 分子名称: | 19-mer fragment of the 23S rRNA, 42-mer fragment of double helix from 16S rRNA, dodecamer fragment of double helix from 23S rRNA | | 著者 | Agrawal, R.K, Sharma, M.R, Kiel, M.C, Hirokawa, G, Booth, T.M, Spahn, C.M, Grassucci, R.A, Kaji, A, Frank, J. | | 登録日 | 2004-04-16 | | 公開日 | 2004-06-15 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (12 Å) | | 主引用文献 | Visualization of ribosome-recycling factor on the Escherichia coli 70S ribosome: Functional implications
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
