3ILM
 
 | | Crystal Structure of the Alr3790 protein from Anabaena sp. Northeast Structural Genomics Consortium Target NsR437h | | Descriptor: | Alr3790 protein, MANGANESE (II) ION | | Authors: | Vorobiev, S, Chen, Y, Maglaqui, M, Ciccosanti, C, Mao, L, Xiao, R, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-08-07 | | Release date: | 2009-09-22 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.262 Å) | | Cite: | Crystal Structure of the Alr3790 protein from Anabaena sp.
To be Published
|
|
3HFQ
 
 | | Crystal structure of the lp_2219 protein from Lactobacillus plantarum. Northeast Structural Genomics Consortium Target LpR118. | | Descriptor: | PHOSPHATE ION, uncharacterized protein lp_2219 | | Authors: | Vorobiev, S.M, Scott, L, Schauder, C, Xiao, R, Ciccosanti, C, Foote, E.L, Maglaqui, M, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-05-12 | | Release date: | 2009-05-26 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (1.963 Å) | | Cite: | Crystal structure of the lp_2219 protein from Lactobacillus plantarum.
To be Published
|
|
3HH2
 
 | |
3FHW
 
 | | Crystal structure of the protein priB from Bordetella parapertussis. Northeast Structural Genomics Consortium target BpR162. | | Descriptor: | DI(HYDROXYETHYL)ETHER, Primosomal replication protein n, SODIUM ION | | Authors: | Kuzin, A.P, Neely, H, Seetharaman, J, Forouhar, F, Wang, D, Mao, L, Maglaqui, M, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-12-10 | | Release date: | 2008-12-30 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the protein priB from Bordetella parapertussis. Northeast Structural Genomics Consortium target BpR162.
To be Published
|
|
3IGN
 
 | | Crystal Structure of the GGDEF domain from Marinobacter aquaeolei diguanylate cyclase complexed with c-di-GMP - Northeast Structural Genomics Consortium Target MqR89a | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Diguanylate cyclase | | Authors: | Vorobiev, S, Neely, H, Seetharaman, J, Wang, H, Foote, E.L, Ciccosanti, C, Sahdev, S, Xiao, R, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-07-28 | | Release date: | 2009-08-11 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal structure of a catalytically active GG(D/E)EF diguanylate cyclase domain from Marinobacter aquaeolei with bound c-di-GMP product.
J.Struct.Funct.Genom., 13, 2012
|
|
3ICL
 
 | | X-Ray Structure of Protein (EAL/GGDEF domain protein) from M.capsulatus, Northeast Structural Genomics Consortium Target McR174C | | Descriptor: | EAL/GGDEF domain protein, SULFATE ION | | Authors: | Kuzin, A, Chen, Y, Seetharaman, J, Mao, M, Xiao, R, Ciccosanti, C, Foote, E.L, Wang, H, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-07-17 | | Release date: | 2009-08-04 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Northeast Structural Genomics Consortium Target McR174C
To be Published
|
|
3EXA
 
 | | Crystal structure of the full-length tRNA isopentenylpyrophosphate transferase (BH2366) from Bacillus halodurans, Northeast Structural Genomics Consortium target BhR41. | | Descriptor: | tRNA delta(2)-isopentenylpyrophosphate transferase | | Authors: | Forouhar, F, Abashidze, M, Neely, H, Seetharaman, J, Shastry, R, Janjua, H, Cunningham, K, Ma, L.-C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-10-16 | | Release date: | 2008-11-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the full-length tRNA isopentenylpyrophosphate transferase (BH2366) from Bacillus halodurans, Northeast Structural Genomics Consortium target BhR41.
To be Published
|
|
3FLH
 
 | | Crystal structure of lp_1913 protein from Lactobacillus plantarum,Northeast Structural Genomics Consortium Target LpR140B | | Descriptor: | BROMIDE ION, uncharacterized protein lp_1913 | | Authors: | Seetharaman, J, Chen, Y, Forouhar, F, Sahdev, S, Foote, E.L, Ciccosanti, C, Belote, R.L, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-12-18 | | Release date: | 2009-01-13 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of lp_1913 protein from Lactobacillus plantarum,Northeast Structural Genomics Consortium Target LpR140B
To be Published
|
|
3F1X
 
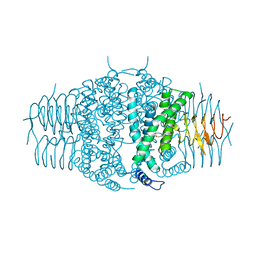 | | Three dimensional structure of the serine acetyltransferase from Bacteroides vulgatus, NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET BVR62. | | Descriptor: | Serine acetyltransferase | | Authors: | Kuzin, A.P, Chen, Y, Seetharaman, J, Sahdev, S, Wang, D, Mao, L, Cunningham, K, Maglaqui, M, Xiao, R, Liu, J, Everett, J.K, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-10-28 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three dimensional structure of the serine acetyltransferase from Bacteroides vulgatus, NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET BVR62.
To be Published
|
|
5OJ4
 
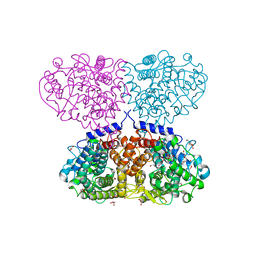 | | Crystal structure of Mycolicibacterium hassiacum glucosylglycerate hydrolase (MhGgH) D182A variant in complex with mannosylglycerate | | Descriptor: | (2R)-3-hydroxy-2-(alpha-D-mannopyranosyloxy)propanoic acid, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Cereija, T.B, Macedo-Ribeiro, S, Pereira, P.J.B. | | Deposit date: | 2017-07-20 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | The structural characterization of a glucosylglycerate hydrolase provides insights into the molecular mechanism of mycobacterial recovery from nitrogen starvation.
Iucrj, 6, 2019
|
|
5OJU
 
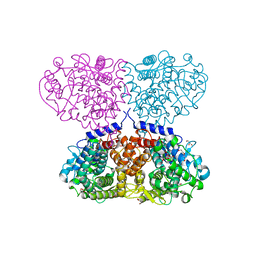 | | Crystal structure of Mycolicibacterium hassiacum glucosylglycerate hydrolase (MhGgH) E419A variant in complex with glucosylglycerate | | Descriptor: | (2R)-2-(alpha-D-glucopyranosyloxy)-3-hydroxypropanoic acid, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Cereija, T.B, Macedo-Ribeiro, S, Pereira, P.J.B. | | Deposit date: | 2017-07-24 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | The structural characterization of a glucosylglycerate hydrolase provides insights into the molecular mechanism of mycobacterial recovery from nitrogen starvation.
Iucrj, 6, 2019
|
|
5OIE
 
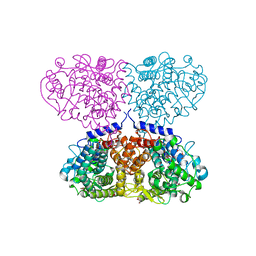 | | Crystal structure of Mycolicibacterium hassiacum glucosylglycerate hydrolase (MhGgH) E419A variant in complex with serine and glycerol | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, SERINE, ... | | Authors: | Cereija, T.B, Macedo-Ribeiro, S, Pereira, P.J.B. | | Deposit date: | 2017-07-18 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.071 Å) | | Cite: | The structural characterization of a glucosylglycerate hydrolase provides insights into the molecular mechanism of mycobacterial recovery from nitrogen starvation.
Iucrj, 6, 2019
|
|
5OIV
 
 | | Crystal structure of Mycolicibacterium hassiacum glucosylglycerate hydrolase (MhGgH) D43A variant in complex with serine and glycerol | | Descriptor: | GLYCEROL, GLYCINE, Hydrolase, ... | | Authors: | Cereija, T.B, Macedo-Ribeiro, S, Pereira, P.J.B. | | Deposit date: | 2017-07-19 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.783 Å) | | Cite: | The structural characterization of a glucosylglycerate hydrolase provides insights into the molecular mechanism of mycobacterial recovery from nitrogen starvation.
Iucrj, 6, 2019
|
|
5ONZ
 
 | | Crystal structure of Mycolicibacterium hassiacum glucosylglycerate hydrolase (MhGgH) D182A variant in complex with glucosylglycolate | | Descriptor: | (alpha-D-glucopyranosyloxy)acetic acid, GLYCEROL, Hydrolase, ... | | Authors: | Cereija, T.B, Macedo-Ribeiro, S, Pereira, P.J.B. | | Deposit date: | 2017-08-04 | | Release date: | 2018-08-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | The structural characterization of a glucosylglycerate hydrolase provides insights into the molecular mechanism of mycobacterial recovery from nitrogen starvation.
Iucrj, 6, 2019
|
|
7SW4
 
 | | MicroED structure of proteinase K from a 540 nm thick lamella measured at 200 kV | | Descriptor: | Proteinase K | | Authors: | Martynowycz, M.W, Clabbers, M.T.B, Unge, J, Hattne, J, Gonen, T. | | Deposit date: | 2021-11-19 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-18 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.4 Å) | | Cite: | Benchmarking the ideal sample thickness in cryo-EM.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7SW9
 
 | | MicroED structure of proteinase K from a 170 nm thick lamella measured at 300 kV | | Descriptor: | Proteinase K | | Authors: | Martynowycz, M.W, Clabbers, M.T.B, Unge, J, Hattne, J, Gonen, T. | | Deposit date: | 2021-11-19 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-18 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.1 Å) | | Cite: | Benchmarking the ideal sample thickness in cryo-EM.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7SVY
 
 | | MicroED structure of proteinase K from a 130 nm thick lamella measured at 120 kV | | Descriptor: | Proteinase K | | Authors: | Martynowycz, M.W, Clabbers, M.T.B, Unge, J, Hattne, J, Gonen, T. | | Deposit date: | 2021-11-19 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-18 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.3 Å) | | Cite: | Benchmarking the ideal sample thickness in cryo-EM.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5ONT
 
 | | Crystal structure of Mycolicibacterium hassiacum glucosylglycerate hydrolase(MhGgH) E419A variant in complex with glucosylglycerol | | Descriptor: | 1,3-dihydroxypropan-2-yl alpha-D-glucopyranoside, GLYCEROL, Uncharacterized protein | | Authors: | Cereija, T.B, Macedo-Ribeiro, S, Pereira, P.J.B. | | Deposit date: | 2017-08-04 | | Release date: | 2018-08-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The structural characterization of a glucosylglycerate hydrolase provides insights into the molecular mechanism of mycobacterial recovery from nitrogen starvation.
Iucrj, 6, 2019
|
|
7SW2
 
 | | MicroED structure of proteinase K from a 130 nm thick lamella measured at 200 kV | | Descriptor: | Proteinase K | | Authors: | Martynowycz, M.W, Clabbers, M.T.B, Unge, J, Hattne, J, Gonen, T. | | Deposit date: | 2021-11-19 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-18 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.95 Å) | | Cite: | Benchmarking the ideal sample thickness in cryo-EM.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7SW8
 
 | | MicroED structure of proteinase K from a 150 nm thick lamella measured at 300 kV | | Descriptor: | Proteinase K | | Authors: | Martynowycz, M.W, Clabbers, M.T.B, Unge, J, Hattne, J, Gonen, T. | | Deposit date: | 2021-11-19 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-18 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.9 Å) | | Cite: | Benchmarking the ideal sample thickness in cryo-EM.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7SWB
 
 | | MicroED structure of proteinase K from a 360 nm thick lamella measured at 300 kV | | Descriptor: | Proteinase K | | Authors: | Martynowycz, M.W, Clabbers, M.T.B, Unge, J, Hattne, J, Gonen, T. | | Deposit date: | 2021-11-19 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-18 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.05 Å) | | Cite: | Benchmarking the ideal sample thickness in cryo-EM.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7SW5
 
 | | MicroED structure of proteinase K from a 460 nm thick lamella measured at 200 kV | | Descriptor: | Proteinase K | | Authors: | Martynowycz, M.W, Clabbers, M.T.B, Unge, J, Hattne, J, Gonen, T. | | Deposit date: | 2021-11-19 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-18 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.95 Å) | | Cite: | Benchmarking the ideal sample thickness in cryo-EM.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7SVZ
 
 | | MicroED structure of proteinase K from a 200 nm thick lamella measured at 120 kV | | Descriptor: | Proteinase K | | Authors: | Martynowycz, M.W, Clabbers, M.T.B, Unge, J, Hattne, J, Gonen, T. | | Deposit date: | 2021-11-19 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-18 | | Method: | ELECTRON CRYSTALLOGRAPHY (2 Å) | | Cite: | Benchmarking the ideal sample thickness in cryo-EM.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7SW0
 
 | | MicroED structure of proteinase K from a 325 nm thick lamella measured at 120 kV | | Descriptor: | Proteinase K | | Authors: | Martynowycz, M.W, Clabbers, M.T.B, Unge, J, Hattne, J, Gonen, T. | | Deposit date: | 2021-11-19 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-18 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.7 Å) | | Cite: | Benchmarking the ideal sample thickness in cryo-EM.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7SW1
 
 | | MicroED structure of proteinase K from a 115 nm thick lamella measured at 200 kV | | Descriptor: | Proteinase K | | Authors: | Martynowycz, M.W, Clabbers, M.T.B, Unge, J, Hattne, J, Gonen, T. | | Deposit date: | 2021-11-19 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-18 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.85 Å) | | Cite: | Benchmarking the ideal sample thickness in cryo-EM.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
