7GRL
 
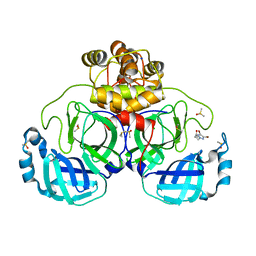 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-8 | | Descriptor: | 3C-like proteinase nsp5, 4-(4,5-dibromo-2H-1,2,3-triazol-2-yl)butan-2-one, CHLORIDE ION, ... | | Authors: | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | Deposit date: | 2023-11-14 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7GRO
 
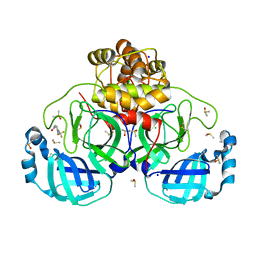 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-11 | | Descriptor: | 3C-like proteinase nsp5, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | Deposit date: | 2023-11-14 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7GRQ
 
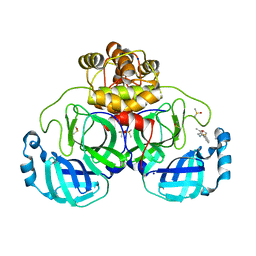 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-13 | | Descriptor: | 3C-like proteinase nsp5, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | Deposit date: | 2023-11-14 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7GRN
 
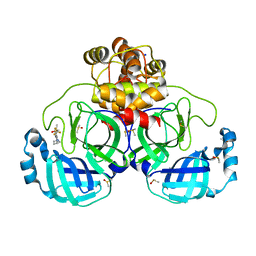 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-10 | | Descriptor: | 2-[(3S)-pyrrolidin-3-yl]-5-(trifluoromethyl)-1H-benzimidazole, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, ... | | Authors: | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | Deposit date: | 2023-11-14 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7GRJ
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-6 | | Descriptor: | (5-chloro-1-benzothiophen-3-yl)methanol, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, ... | | Authors: | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | Deposit date: | 2023-11-14 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7GRW
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-19 | | Descriptor: | (2S)-N-(3,5-dichlorophenyl)-2-hydroxypropanamide, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, ... | | Authors: | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | Deposit date: | 2023-11-14 | | Release date: | 2024-02-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7GRX
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-20 | | Descriptor: | 1-(2,4-difluorophenyl)pyrrolidine-2,5-dione, 3C-like proteinase nsp5, CHLORIDE ION, ... | | Authors: | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | Deposit date: | 2023-11-14 | | Release date: | 2024-02-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7GRS
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-15 | | Descriptor: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, SODIUM ION, ... | | Authors: | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | Deposit date: | 2023-11-14 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7GRM
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-9 | | Descriptor: | 3C-like proteinase nsp5, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | Deposit date: | 2023-11-14 | | Release date: | 2024-02-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7GRV
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-18 | | Descriptor: | (2S)-2-(2-fluorophenyl)-1,3-thiazolidin-4-one, 3C-like proteinase nsp5, CHLORIDE ION, ... | | Authors: | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | Deposit date: | 2023-11-14 | | Release date: | 2024-02-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7GRU
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-17 | | Descriptor: | 3-(4-chlorophenyl)-1-methyl-1H-pyrazol-5-amine, 3C-like proteinase nsp5, CHLORIDE ION, ... | | Authors: | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | Deposit date: | 2023-11-14 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7GS1
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-24 | | Descriptor: | 2-cyano-~{N}-cyclohexyl-ethanamide, 3C-like proteinase nsp5, CHLORIDE ION, ... | | Authors: | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | Deposit date: | 2023-11-14 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7GS0
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-23 | | Descriptor: | (pyridin-2-yl)(quinolin-2-yl)methanone, 3C-like proteinase nsp5, CHLORIDE ION, ... | | Authors: | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | Deposit date: | 2023-11-14 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
5NB7
 
 | | Complement factor D | | Descriptor: | 1-[2-[(1~{R},3~{S},5~{R})-3-[(6-bromanylpyridin-2-yl)carbamoyl]-2-azabicyclo[3.1.0]hexan-2-yl]-2-oxidanylidene-ethyl]indazole-3-carboxamide, Complement factor D, DIMETHYL SULFOXIDE | | Authors: | Mac Sweeney, A, Ostermann, N. | | Deposit date: | 2017-03-01 | | Release date: | 2017-06-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Discovery of Highly Potent and Selective Small-Molecule Reversible Factor D Inhibitors Demonstrating Alternative Complement Pathway Inhibition in Vivo.
J. Med. Chem., 60, 2017
|
|
6FUH
 
 | | Complement factor D in complex with the inhibitor (4-((3-(aminomethyl)phenyl)amino)quinazolin-2-yl)-L-valine | | Descriptor: | (2~{S})-2-[[4-[[3-(aminomethyl)phenyl]amino]quinazolin-2-yl]amino]-3-methyl-butanoic acid, Complement factor D | | Authors: | Mac Sweeney, A, Ostermann, N, Vulpetti, A, Maibaum, J, Erbel, P, Lorthiois, E, Yoon, T, Randl, S, Ruedisser, S. | | Deposit date: | 2018-02-27 | | Release date: | 2018-06-06 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Discovery and Design of First Benzylamine-Based Ligands Binding to an Unlocked Conformation of the Complement Factor D.
ACS Med Chem Lett, 9, 2018
|
|
5NAR
 
 | |
5NAW
 
 | | Complement factor D in complex with the inhibitor (1R,3S,5R)-2-Aza-bicyclo[3.1.0]hexane-2,3-dicarboxylic acid 2-[(1-carbamoyl-1H-indol-3-yl)-amide] 3-[(3-trifluoromethoxy-phenyl)-amide] | | Descriptor: | (1~{R},3~{S},5~{R})-~{N}2-(1-aminocarbonylindol-3-yl)-~{N}3-[3-(trifluoromethyloxy)phenyl]-2-azabicyclo[3.1.0]hexane-2,3-dicarboxamide, Complement factor D | | Authors: | Mac Sweeney, A, Ostermann, N. | | Deposit date: | 2017-02-28 | | Release date: | 2017-06-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Discovery of Highly Potent and Selective Small-Molecule Reversible Factor D Inhibitors Demonstrating Alternative Complement Pathway Inhibition in Vivo.
J. Med. Chem., 60, 2017
|
|
6FUI
 
 | | Complement factor D in complex with the inhibitor 3-((3-((3-(aminomethyl)phenyl)amino)-1H-pyrazolo[3,4-d]pyrimidin-4-yl)amino)phenol | | Descriptor: | (1~{R},2~{S})-2-[[4-[[3-(aminomethyl)phenyl]amino]quinazolin-2-yl]amino]cyclohexane-1-carboxylic acid, Complement factor D | | Authors: | Mac Sweeney, A, Ostermann, N, Vulpetti, A, Maibaum, J, Erbel, P, Lorthiois, E, Yoon, T, Randl, S, Ruedisser, S. | | Deposit date: | 2018-02-27 | | Release date: | 2018-06-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Discovery and Design of First Benzylamine-Based Ligands Binding to an Unlocked Conformation of the Complement Factor D.
ACS Med Chem Lett, 9, 2018
|
|
5NAT
 
 | |
6FUG
 
 | | Complement factor D in complex with the inhibitor 3-((3-((3-(aminomethyl)phenyl)amino)-1H-pyrazolo[3,4-d]pyrimidin-4-yl)amino)phenol | | Descriptor: | 3-[[3-[[3-(aminomethyl)phenyl]amino]-1~{H}-pyrazolo[3,4-d]pyrimidin-4-yl]amino]phenol, Complement factor D | | Authors: | Mac Sweeney, A, Ostermann, N, Vulpetti, A, Maibaum, J, Erbel, P, Lorthiois, E, Yoon, T, Randl, S, Ruedisser, S. | | Deposit date: | 2018-02-27 | | Release date: | 2018-06-06 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Discovery and Design of First Benzylamine-Based Ligands Binding to an Unlocked Conformation of the Complement Factor D.
ACS Med Chem Lett, 9, 2018
|
|
6FUT
 
 | | Complement factor D in complex with the inhibitor (S)-3'-(aminomethyl)-N-(1,2,3,4-tetrahydronaphthalen-1-yl)-[1,1'-biphenyl]-3-carboxamide | | Descriptor: | 3-[3-(aminomethyl)phenyl]-~{N}-[(1~{S})-1,2,3,4-tetrahydronaphthalen-1-yl]benzamide, Complement factor D, SUCCINIC ACID | | Authors: | Mac Sweeney, A, Vulpetti, A, Erbel, P, Lorthiois, E, Maibaum, J, Randl, S. | | Deposit date: | 2018-02-27 | | Release date: | 2018-06-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Discovery and Design of First Benzylamine-Based Ligands Binding to an Unlocked Conformation of the Complement Factor D.
ACS Med Chem Lett, 9, 2018
|
|
5NB6
 
 | | Complement factor D in complex with the inhibitor (2S,4S)-4-Amino-pyrrolidine-1,2-dicarboxylic acid 1-[(1-carbamoyl-1H-indol-3-yl)-amide] 2-[(3-trifluoromethoxy-phenyl)-amide] | | Descriptor: | (2~{S},4~{S})-~{N}1-(1-aminocarbonylindol-3-yl)-4-azanyl-~{N}2-[3-(trifluoromethyloxy)phenyl]pyrrolidine-1,2-dicarboxamide, Complement factor D | | Authors: | Mac Sweeney, A, Ostermann, N. | | Deposit date: | 2017-03-01 | | Release date: | 2017-06-28 | | Last modified: | 2017-07-26 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Discovery of Highly Potent and Selective Small-Molecule Reversible Factor D Inhibitors Demonstrating Alternative Complement Pathway Inhibition in Vivo.
J. Med. Chem., 60, 2017
|
|
5NBA
 
 | | Complement factor D in complex with the inhibitor (2S,4R)-4-Fluoro-pyrrolidine-1,2-dicarboxylic acid 1-[(1-carbamoyl-1H-indol-3-yl)-amide] 2-[(3-trifluoromethoxy-phenyl)-amide] | | Descriptor: | (2~{S},4~{R})-~{N}1-(1-aminocarbonylindol-3-yl)-4-fluoranyl-~{N}2-[3-(trifluoromethyloxy)phenyl]pyrrolidine-1,2-dicarboxamide, Complement factor D | | Authors: | Mac Sweeney, A, Ostermann, N. | | Deposit date: | 2017-03-01 | | Release date: | 2017-06-28 | | Last modified: | 2017-07-26 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Discovery of Highly Potent and Selective Small-Molecule Reversible Factor D Inhibitors Demonstrating Alternative Complement Pathway Inhibition in Vivo.
J. Med. Chem., 60, 2017
|
|
6FUJ
 
 | | Complement factor D in complex with the inhibitor N-(3'-(aminomethyl)-[1,1'-biphenyl]-3-yl)-3-methylbutanamide | | Descriptor: | Complement factor D, ~{N}-[3-[3-(aminomethyl)phenyl]phenyl]-3-methyl-butanamide | | Authors: | Mac Sweeney, A, Ostermann, N, Vulpetti, A, Maibaum, J, Erbel, P, Lorthiois, E, Yoon, T, Randl, S, Ruedisser, S. | | Deposit date: | 2018-02-27 | | Release date: | 2018-06-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Discovery and Design of First Benzylamine-Based Ligands Binding to an Unlocked Conformation of the Complement Factor D.
ACS Med Chem Lett, 9, 2018
|
|
6TA4
 
 | | Human kinesin-5 motor domain in the AMPPNP state bound to microtubules | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Kinesin-like protein KIF11, MAGNESIUM ION, ... | | Authors: | Pena, A.P, Sweeney, A, Cook, A.D, Moores, C.A, Topf, M. | | Deposit date: | 2019-10-29 | | Release date: | 2020-03-04 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Structure of Microtubule-Trapped Human Kinesin-5 and Its Mechanism of Inhibition Revealed Using Cryoelectron Microscopy.
Structure, 28, 2020
|
|
