6N9A
 
 | |
1PYB
 
 | | Crystal Structure of Aquifex aeolicus Trbp111: a Structure-Specific tRNA Binding Protein | | Descriptor: | tRNA-binding protein Trbp111 | | Authors: | Swairjo, M.A, Morales, A.J, Wang, C.C, Ortiz, A.R, Schimmel, P. | | Deposit date: | 2003-07-08 | | Release date: | 2003-08-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of trbp111: a structure-specific tRNA-binding protein.
Embo J., 19, 2000
|
|
1YFR
 
 | |
1YFS
 
 | |
1YGB
 
 | |
1YFT
 
 | |
1RIQ
 
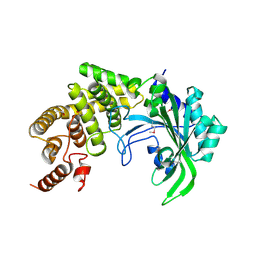 | | The crystal structure of the catalytic fragment of the alanyl-tRNA synthetase | | Descriptor: | Alanyl-tRNA synthetase | | Authors: | Swairjo, M.A, Otero, F.J, Yang, X.-L, Lovato, M.A, Skene, R.J, McRee, D.E, Ribas de Pouplana, L, Schimmel, P. | | Deposit date: | 2003-11-17 | | Release date: | 2004-04-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Alanyl-tRNA Synthetase Crystal Structure and Design for Acceptor-Stem Recognition
Mol.Cell, 13, 2004
|
|
1A8A
 
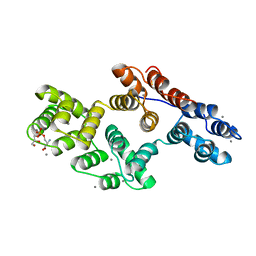 | | RAT ANNEXIN V COMPLEXED WITH GLYCEROPHOSPHOSERINE | | Descriptor: | ANNEXIN V, CALCIUM ION, L-ALPHA-GLYCEROPHOSPHORYLSERINE | | Authors: | Swairjo, M.A, Concha, N.O, Kaetzel, M.A, Dedman, J.R, Seaton, B.A. | | Deposit date: | 1998-03-23 | | Release date: | 1998-06-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ca(2+)-bridging mechanism and phospholipid head group recognition in the membrane-binding protein annexin V.
Nat.Struct.Biol., 2, 1995
|
|
1A8B
 
 | | RAT ANNEXIN V COMPLEXED WITH GLYCEROPHOSPHOETHANOLAMINE | | Descriptor: | ANNEXIN V, CALCIUM ION, L-ALPHA-GLYCEROPHOSPHORYLETHANOLAMINE | | Authors: | Swairjo, M.A, Concha, N.O, Kaetzel, M.A, Dedman, J.R, Seaton, B.A. | | Deposit date: | 1998-03-23 | | Release date: | 1998-06-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ca(2+)-bridging mechanism and phospholipid head group recognition in the membrane-binding protein annexin V.
Nat.Struct.Biol., 2, 1995
|
|
1BDR
 
 | | HIV-1 (2: 31, 33-37) PROTEASE COMPLEXED WITH INHIBITOR SB203386 | | Descriptor: | (2R,4S,5S,1'S)-2-PHENYLMETHYL-4-HYDROXY-5-(TERT-BUTOXYCARBONYL)AMINO-6-PHENYL HEXANOYL-N-(1'-IMIDAZO-2-YL)-2'-METHYLPROPANAMIDE, HIV-1 PROTEASE | | Authors: | Swairjo, M.A, Abdel-Meguid, S.S. | | Deposit date: | 1998-05-10 | | Release date: | 1998-10-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural role of the 30's loop in determining the ligand specificity of the human immunodeficiency virus protease.
Biochemistry, 37, 1998
|
|
1BDQ
 
 | | HIV-1 (2:31-37, 47, 82) PROTEASE COMPLEXED WITH INHIBITOR SB203386 | | Descriptor: | (2R,4S,5S,1'S)-2-PHENYLMETHYL-4-HYDROXY-5-(TERT-BUTOXYCARBONYL)AMINO-6-PHENYL HEXANOYL-N-(1'-IMIDAZO-2-YL)-2'-METHYLPROPANAMIDE, HIV-1 PROTEASE | | Authors: | Swairjo, M.A, Abdel-Meguid, S.S. | | Deposit date: | 1998-05-10 | | Release date: | 1998-08-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural role of the 30's loop in determining the ligand specificity of the human immunodeficiency virus protease.
Biochemistry, 37, 1998
|
|
1BDL
 
 | | HIV-1 (2:31-37) PROTEASE COMPLEXED WITH INHIBITOR SB203386 | | Descriptor: | (2R,4S,5S,1'S)-2-PHENYLMETHYL-4-HYDROXY-5-(TERT-BUTOXYCARBONYL)AMINO-6-PHENYL HEXANOYL-N-(1'-IMIDAZO-2-YL)-2'-METHYLPROPANAMIDE, HIV-1 PROTEASE | | Authors: | Swairjo, M.A, Abdel-Meguid, S.S. | | Deposit date: | 1998-05-10 | | Release date: | 1998-08-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural role of the 30's loop in determining the ligand specificity of the human immunodeficiency virus protease.
Biochemistry, 37, 1998
|
|
3D2O
 
 | | Crystal Structure of Manganese-metallated GTP Cyclohydrolase Type IB | | Descriptor: | AZIDE ION, CHLORIDE ION, LITHIUM ION, ... | | Authors: | Swairjo, M.A. | | Deposit date: | 2008-05-08 | | Release date: | 2009-05-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Zinc-independent folate biosynthesis: genetic, biochemical, and structural investigations reveal new metal dependence for GTP cyclohydrolase IB
J.Bacteriol., 191, 2009
|
|
3ERS
 
 | | Crystal Structure of E. coli Trbp111 | | Descriptor: | tRNA-binding protein ygjH | | Authors: | Swairjo, M.A. | | Deposit date: | 2008-10-03 | | Release date: | 2008-12-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal structure of Trbp111: a tructure specific tRNA binding protein
Embo J., 19, 2000
|
|
8DL3
 
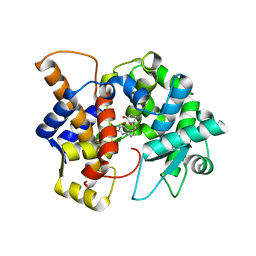 | | Crystal structure of the human queuine salvage enzyme DUF2419, complexed with queuine | | Descriptor: | 2-amino-5-({[(1S,4S,5R)-4,5-dihydroxycyclopent-2-en-1-yl]amino}methyl)-3,7-dihydro-4H-pyrrolo[2,3-d]pyrimidin-4-one, Queuosine salvage protein | | Authors: | Hung, S.-H, Swairjo, M.A. | | Deposit date: | 2022-07-06 | | Release date: | 2022-12-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structural basis of Qng1-mediated salvage of the micronutrient queuine from queuosine-5'-monophosphate as the biological substrate.
Nucleic Acids Res., 51, 2023
|
|
2AZX
 
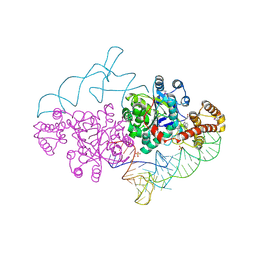 | | Charged and uncharged tRNAs adopt distinct conformations when complexed with human tryptophanyl-tRNA synthetase | | Descriptor: | 72-MER, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Yang, X.L, Otero, F.J, Ewalt, K.L, Liu, J, Swairjo, M.A, Kohrer, C, RajBhandary, U.L, Skene, R.J, McRee, D.E, Schimmel, P. | | Deposit date: | 2005-09-12 | | Release date: | 2006-08-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Two conformations of a crystalline human tRNA synthetase-tRNA complex: implications for protein synthesis.
Embo J., 25, 2006
|
|
5K9G
 
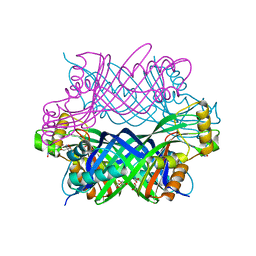 | | Crystal Structure of GTP Cyclohydrolase-IB with Tris | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FORMIC ACID, ... | | Authors: | Alvarez, J, Stec, B, Swairjo, M.A. | | Deposit date: | 2016-05-31 | | Release date: | 2016-09-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanism and catalytic strategy of the prokaryotic-specific GTP cyclohydrolase-IB.
Biochem.J., 474, 2017
|
|
5K95
 
 | |
5JYX
 
 | |
5K0P
 
 | |
7U07
 
 | |
7U1O
 
 | | Crystal structure of queuine salvage enzyme DUF2419 complexed with queuosine | | Descriptor: | 2-amino-5-({[(1S,4S,5R)-4,5-dihydroxycyclopent-2-en-1-yl]amino}methyl)-7-beta-D-ribofuranosyl-3,7-dihydro-4H-pyrrolo[2,3-d]pyrimidin-4-one, DI(HYDROXYETHYL)ETHER, MALONATE ION, ... | | Authors: | Hung, S.-H, Swairjo, M.A. | | Deposit date: | 2022-02-21 | | Release date: | 2022-12-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis of Qng1-mediated salvage of the micronutrient queuine from queuosine-5'-monophosphate as the biological substrate.
Nucleic Acids Res., 51, 2023
|
|
7U5A
 
 | | Crystal structure of queuine salvage enzyme DUF2419 mutant K199C, complexed with queuosine | | Descriptor: | 2-amino-5-({[(1S,4S,5R)-4,5-dihydroxycyclopent-2-en-1-yl]amino}methyl)-7-beta-D-ribofuranosyl-3,7-dihydro-4H-pyrrolo[2,3-d]pyrimidin-4-one, MALONATE ION, Queuine salvage enzyme DUF2419 | | Authors: | Hung, S.-H, Swairjo, M.A. | | Deposit date: | 2022-03-01 | | Release date: | 2022-12-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of Qng1-mediated salvage of the micronutrient queuine from queuosine-5'-monophosphate as the biological substrate.
Nucleic Acids Res., 51, 2023
|
|
7UK3
 
 | |
7U91
 
 | | Crystal structure of queuine salvage enzyme DUF2419, in complex with queuosine-5'-monophosphate | | Descriptor: | 2-amino-5-({[(1S,4S,5R)-4,5-dihydroxycyclopent-2-en-1-yl]amino}methyl)-7-(5-O-phosphono-beta-D-ribofuranosyl)-3,7-dihydro-4H-pyrrolo[2,3-d]pyrimidin-4-one, AMMONIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Hung, S.-H, Swairjo, M.A. | | Deposit date: | 2022-03-09 | | Release date: | 2022-12-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of Qng1-mediated salvage of the micronutrient queuine from queuosine-5'-monophosphate as the biological substrate.
Nucleic Acids Res., 51, 2023
|
|
