8HGM
 
 | | Structure of SARS-CoV-2 spike RBD in complex with neutralizing antibody NIV-11 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, NIV-11 Fab heavy chain, NIV-11 Fab light chain, ... | | Authors: | Moriyama, S, Anraku, Y, Muranishi, S, Adachi, Y, Kuroda, D, Higuchi, Y, Kotaki, R, Tonouchi, K, Yumoto, K, Suzuki, T, Kita, S, Someya, T, Fukuhara, H, Kuroda, Y, Yamamoto, T, Onodera, T, Fukushi, S, Maeda, K, Nakamura-Uchiyama, F, Hashiguchi, T, Hoshino, A, Maenaka, K, Takahashi, Y. | | Deposit date: | 2022-11-15 | | Release date: | 2023-10-25 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural delineation and computational design of SARS-CoV-2-neutralizing antibodies against Omicron subvariants.
Nat Commun, 14, 2023
|
|
8HGL
 
 | | SARS-CoV-2 spike in complex with neutralizing antibody NIV-11 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, NIV-11 Fab heavy chain, ... | | Authors: | Moriyama, S, Anraku, Y, Muranishi, S, Adachi, Y, Kuroda, D, Higuchi, Y, Kotaki, R, Tonouchi, K, Yumoto, K, Suzuki, T, Kita, S, Someya, T, Fukuhara, H, Kuroda, Y, Yamamoto, T, Onodera, T, Fukushi, S, Maeda, K, Nakamura-Uchiyama, F, Hashiguchi, T, Hoshino, A, Maenaka, K, Takahashi, Y. | | Deposit date: | 2022-11-15 | | Release date: | 2023-10-25 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural delineation and computational design of SARS-CoV-2-neutralizing antibodies against Omicron subvariants.
Nat Commun, 14, 2023
|
|
8HES
 
 | | Crystal structure of SARS-CoV-2 RBD and NIV-10 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, NIV-10 Fab H-chain, NIV-10 Fab L-chain, ... | | Authors: | Moriyama, S, Anraku, Y, Taminishi, S, Adachi, Y, Kuroda, D, Higuchi, Y, Kotaki, R, Tonouchi, K, Yumoto, K, Suzuki, T, Kita, S, Someya, T, Fukuhara, H, Kuroda, Y, Yamamoto, T, Onodera, T, Fukushi, S, Maeda, K, Nakamura-Uchiyama, F, Hashiguchi, T, Hoshino, A, Maenaka, K, Takahashi, Y. | | Deposit date: | 2022-11-08 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural delineation and computational design of SARS-CoV-2-neutralizing antibodies against Omicron subvariants.
Nat Commun, 14, 2023
|
|
4K7B
 
 | | Crystal structure of Extrinsic protein in photosystem II | | Descriptor: | Extrinsic protein in photosystem II, GLYCEROL, SULFATE ION | | Authors: | Nagao, R, Suga, M, Niikura, A, Okumura, A, Koua, F.H.M, Suzuki, T, Tomo, T, Enami, I, Shen, J.R. | | Deposit date: | 2013-04-16 | | Release date: | 2013-09-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal Structure of Psb31, a Novel Extrinsic Protein of Photosystem II from a Marine Centric Diatom and Implications for Its Binding and Function
Biochemistry, 52, 2013
|
|
7WEM
 
 | | Solid-state NMR Structure of TFo c-Subunit Ring | | Descriptor: | ATP synthase subunit c | | Authors: | Akutsu, H, Todokoro, Y, Kang, S.-J, Suzuki, T, Yoshida, M, Ikegami, T, Fujiwara, T. | | Deposit date: | 2021-12-23 | | Release date: | 2022-08-10 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Chemical Conformation of the Essential Glutamate Site of the c -Ring within Thermophilic Bacillus F o F 1 -ATP Synthase Determined by Solid-State NMR Based on its Isolated c -Ring Structure.
J.Am.Chem.Soc., 144, 2022
|
|
2E4M
 
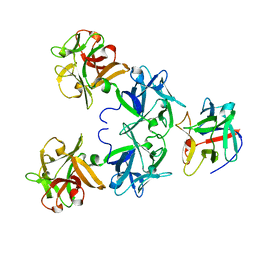 | | Crystal structure of hemagglutinin subcomponent complex (HA-33/HA-17) from Clostridium botulinum serotype D strain 4947 | | Descriptor: | HA-17, Main hemagglutinin component | | Authors: | Hasegawa, K, Watanabe, T, Suzuki, T, Yamano, A, Niwa, K, Ohyama, T. | | Deposit date: | 2006-12-13 | | Release date: | 2007-06-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A Novel Subunit Structure of Clostridium botulinum Serotype D Toxin Complex with Three Extended Arms
J.Biol.Chem., 282, 2007
|
|
7VNW
 
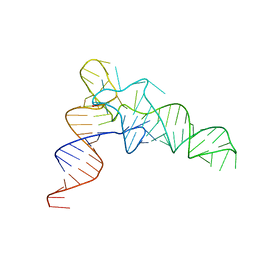 | |
7VNV
 
 | |
7VNX
 
 | | Crystal structure of TkArkI | | Descriptor: | GUANOSINE, TkArkI | | Authors: | Yamashita, S, Minowa, K, Ohira, T, Suzuki, T, Tomita, K. | | Deposit date: | 2021-10-12 | | Release date: | 2022-05-04 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Reversible RNA phosphorylation stabilizes tRNA for cellular thermotolerance.
Nature, 605, 2022
|
|
5EO8
 
 | | Crystal structure of AOL(868) | | Descriptor: | Predicted protein, methyl 1-seleno-beta-L-fucopyranoside | | Authors: | Kato, R, Kiso, M, Ishida, H, Ando, H, Suzuki, T, Shimabukuro, S, Makyio, H. | | Deposit date: | 2015-11-10 | | Release date: | 2016-06-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Six independent fucose-binding sites in the crystal structure of Aspergillus oryzae lectin
Biochem.Biophys.Res.Commun., 477, 2016
|
|
7BPG
 
 | | Structure of serinol nucleic acid - RNA complex | | Descriptor: | CALCIUM ION, RNA (5'-R(*GP*CP*UP*GP*CP*(5BU)P*GP*C)-3'), SNA (S-(F7R)(F7X)(F7O)(F7R)(F7X)(F7O)(F7R)(F7U)-R) | | Authors: | Kamiya, Y, Satoh, T, Kodama, A, Suzuki, T, Uchiyama, S, Kato, K, Asanuma, H. | | Deposit date: | 2020-03-22 | | Release date: | 2020-11-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Intrastrand backbone-nucleobase interactions stabilize unwound right-handed helical structures of heteroduplexes of L-aTNA/RNA and SNA/RNA
Commun Chem, 2020
|
|
2UXD
 
 | | Crystal structure of an extended tRNA anticodon stem loop in complex with its cognate mRNA CGGG in the context of the Thermus thermophilus 30S subunit. | | Descriptor: | 16S RIBOSOMAL RNA, A-SITE MESSENGER RNA FRAGMENT CGGG, ANTICODON STEM-LOOP OF TRANSFER RNA WITH ANTICODON CCCG, ... | | Authors: | Dunham, C.M, Selmer, M, Phelps, S.S, Kelley, A.C, Suzuki, T, Joseph, S, Ramakrishnan, V. | | Deposit date: | 2007-03-28 | | Release date: | 2007-10-02 | | Last modified: | 2019-10-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structures of Trnas with an Expanded Anticodon Loop in the Decoding Center of the 30S Ribosomal Subunit.
RNA, 13, 2007
|
|
2UXB
 
 | | Crystal structure of an extended tRNA anticodon stem loop in complex with its cognate mRNA GGGU in the context of the Thermus thermophilus 30S subunit. | | Descriptor: | 16S RIBOSOMAL RNA, A-SITE MESSENGER RNA FRAGMENT GGGU, ANTICODON STEM-LOOP OF TRANSFER RNA WITH ANTICODON ACCC, ... | | Authors: | Dunham, C.M, Selmer, M, Phelps, S.S, Kelley, A.C, Suzuki, T, Joseph, S, Ramakrishnan, V. | | Deposit date: | 2007-03-28 | | Release date: | 2007-07-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of tRNAs with an expanded anticodon loop in the decoding center of the 30S ribosomal subunit.
RNA, 13, 2007
|
|
2UXC
 
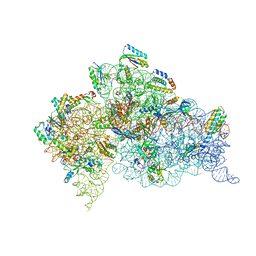 | | Crystal structure of an extended tRNA anticodon stem loop in complex with its cognate mRNA UCGU in the context of the Thermus thermophilus 30S subunit. | | Descriptor: | 16S RIBOSOMAL RNA, A-SITE MESSENGER RNA FRAGMENT CGGG, ANTICODON STEM-LOOP OF TRANSFER RNA WITH ANTICODON ACGA, ... | | Authors: | Dunham, C.M, Selmer, M, Phelps, S.S, Kelley, A.C, Suzuki, T, Joseph, S, Ramakrishnan, V. | | Deposit date: | 2007-03-28 | | Release date: | 2007-05-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of Trnas with an Expanded Anticodon Loop in the Decoding Center of the 30S Ribosomal Subunit.
RNA, 13, 2007
|
|
7BPF
 
 | | Structure of L-threoninol nucleic acid - RNA complex | | Descriptor: | L-aTNA (3'-(*GP*CP*AP*GP*CP*AP*GP*C)-1'), RNA (5'-R(*GP*CP*UP*GP*CP*(5BU)P*GP*C)-3') | | Authors: | Kamiya, Y, Satoh, T, Kodama, A, Suzuki, T, Uchiyama, S, Kato, K, Asanuma, H. | | Deposit date: | 2020-03-22 | | Release date: | 2020-11-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Intrastrand backbone-nucleobase interactions stabilize unwound right-handed helical structures of heteroduplexes of L-aTNA/RNA and SNA/RNA
Commun Chem, 2020
|
|
2D1P
 
 | | crystal structure of heterohexameric TusBCD proteins, which are crucial for the tRNA modification | | Descriptor: | Hypothetical UPF0116 protein yheM, Hypothetical UPF0163 protein yheN, Hypothetical protein yheL, ... | | Authors: | Numata, T, Fukai, S, Ikeuchi, Y, Suzuki, T, Nureki, O. | | Deposit date: | 2005-08-30 | | Release date: | 2006-02-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural Basis for Sulfur Relay to RNA Mediated by Heterohexameric TusBCD Complex
Structure, 14, 2006
|
|
2D5U
 
 | |
2DER
 
 | | Cocrystal structure of an RNA sulfuration enzyme MnmA and tRNA-Glu in the initial tRNA binding state | | Descriptor: | PHOSPHATE ION, SULFATE ION, tRNA, ... | | Authors: | Numata, T, Ikeuchi, Y, Fukai, S, Suzuki, T, Nureki, O. | | Deposit date: | 2006-02-17 | | Release date: | 2006-08-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Snapshots of tRNA sulphuration via an adenylated intermediate
Nature, 442, 2006
|
|
2DET
 
 | | Cocrystal structure of an RNA sulfuration enzyme MnmA and tRNA-Glu in the pre-reaction state | | Descriptor: | SULFATE ION, tRNA, tRNA-specific 2-thiouridylase mnmA | | Authors: | Numata, T, Ikeuchi, Y, Fukai, S, Suzuki, T, Nureki, O. | | Deposit date: | 2006-02-17 | | Release date: | 2006-08-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Snapshots of tRNA sulphuration via an adenylated intermediate
Nature, 442, 2006
|
|
2DEU
 
 | | Cocrystal structure of an RNA sulfuration enzyme MnmA and tRNA-Glu in the adenylated intermediate state | | Descriptor: | ADENOSINE MONOPHOSPHATE, SULFATE ION, tRNA, ... | | Authors: | Numata, T, Ikeuchi, Y, Fukai, S, Suzuki, T, Nureki, O. | | Deposit date: | 2006-02-17 | | Release date: | 2006-08-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Snapshots of tRNA sulphuration via an adenylated intermediate
Nature, 442, 2006
|
|
1WY5
 
 | | Crystal structure of isoluecyl-tRNA lysidine synthetase | | Descriptor: | Hypothetical UPF0072 protein AQ_1887 | | Authors: | Nakanishi, K, Fukai, S, Ikeuchi, Y, Soma, A, Sekine, Y, Suzuki, T, Nureki, O, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-02-06 | | Release date: | 2005-05-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structural basis for lysidine formation by ATP pyrophosphatase accompanied by a lysine-specific loop and a tRNA-recognition domain.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1WTH
 
 | | Crystal structure of gp5-S351L mutant and gp27 complex | | Descriptor: | Baseplate structural protein Gp27, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Kanamaru, S, Ishiwata, Y, Suzuki, T, Rossmann, M.G, Arisaka, F. | | Deposit date: | 2004-11-23 | | Release date: | 2005-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Control of bacteriophage t4 tail lysozyme activity during the infection process
J.Mol.Biol., 346, 2005
|
|
4ZY3
 
 | | Crystal Structure of Keap1 in Complex with a small chemical compound, K67 | | Descriptor: | FORMIC ACID, Kelch-like ECH-associated protein 1, N,N'-[2-(2-oxopropyl)naphthalene-1,4-diyl]bis(4-ethoxybenzenesulfonamide) | | Authors: | Fukutomi, T, Iso, T, Suzuki, T, Takagi, K, Mizushima, T, Komatsu, M, Yamamoto, M. | | Deposit date: | 2015-05-21 | | Release date: | 2016-05-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | p62/Sqstm1 promotes malignancy of HCV-positive hepatocellular carcinoma through Nrf2-dependent metabolic reprogramming
Nat Commun, 7, 2016
|
|
6JJN
 
 | | Crystal structure of Mumps virus hemagglutinin-neuraminidase bound to sialyl lewisX | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HN protein, ... | | Authors: | Kubota, M, Matsuoka, R, Suzuki, T, Yonekura, K, Yanagi, Y, Hashiguchi, T. | | Deposit date: | 2019-02-26 | | Release date: | 2019-05-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular Mechanism of the Flexible Glycan Receptor Recognition by Mumps Virus.
J.Virol., 93, 2019
|
|
6JJM
 
 | | Crystal structure of Mumps virus hemagglutinin-neuraminidase bound to the oligosaccharide portion of the GM2 ganglioside | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HN protein, ... | | Authors: | Kubota, M, Matsuoka, R, Suzuki, T, Yonekura, K, Yanagi, Y, Hashiguchi, T. | | Deposit date: | 2019-02-26 | | Release date: | 2019-05-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.049 Å) | | Cite: | Molecular Mechanism of the Flexible Glycan Receptor Recognition by Mumps Virus.
J.Virol., 93, 2019
|
|
