1BQ5
 
 | | NITRITE REDUCTASE FROM ALCALIGENES XYLOSOXIDANS GIFU 1051 | | Descriptor: | COPPER (II) ION, NITRITE REDUCTASE | | Authors: | Inoue, T, Gotowda, M, Deligeer, Suzuki, S, Kataoka, K, Yamaguchi, K, Watanabe, H, Goho, M, Yasushi, K.A.I. | | Deposit date: | 1998-08-21 | | Release date: | 1999-08-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Type 1 Cu structure of blue nitrite reductase from Alcaligenes xylosoxidans GIFU 1051 at 2.05 A resolution: comparison of blue and green nitrite reductases.
J.Biochem.(Tokyo), 124, 1998
|
|
7X9U
 
 | | Type-II KH motif of human mitochondrial RbfA | | Descriptor: | Putative ribosome-binding factor A, mitochondrial | | Authors: | Kuwasako, K, Suzuki, S, Furue, M, Takizawa, M, Takahashi, M, Tsuda, K, Nagata, T, Watanabe, S, Tanaka, A, Kobayashi, N, Kigawa, T, Guntert, P, Shirouzu, M, Yokoyama, S, Muto, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2022-03-16 | | Release date: | 2023-01-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | 1 H, 13 C, and 15 N resonance assignments and solution structures of the KH domain of human ribosome binding factor A, mtRbfA, involved in mitochondrial ribosome biogenesis.
Biomol.Nmr Assign., 16, 2022
|
|
1PMY
 
 | | REFINED CRYSTAL STRUCTURE OF PSEUDOAZURIN FROM METHYLOBACTERIUM EXTORQUENS AM1 AT 1.5 ANGSTROMS RESOLUTION | | Descriptor: | COPPER (II) ION, PSEUDOAZURIN | | Authors: | Inoue, T, Kai, Y, Harada, S, Kasai, N, Ohshiro, Y, Suzuki, S, Kohzuma, T, Tobari, J. | | Deposit date: | 1994-01-28 | | Release date: | 1994-07-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Refined crystal structure of pseudoazurin from Methylobacterium extorquens AM1 at 1.5 A resolution.
Acta Crystallogr.,Sect.D, 50, 1994
|
|
3EF4
 
 | | Crystal structure of native pseudoazurin from Hyphomicrobium denitrificans | | Descriptor: | Blue copper protein, COPPER (II) ION, PHOSPHATE ION | | Authors: | Hira, D, Nojiri, M, Suzuki, S. | | Deposit date: | 2008-09-08 | | Release date: | 2008-12-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Atomic resolution structure of pseudoazurin from the methylotrophic denitrifying bacterium Hyphomicrobium denitrificans: structural insights into its spectroscopic properties
Acta Crystallogr.,Sect.D, 65, 2009
|
|
1CUO
 
 | | CRYSTAL STRUCTURE ANALYSIS OF ISOMER-2 AZURIN FROM METHYLOMONAS J | | Descriptor: | COPPER (II) ION, PROTEIN (AZURIN ISO-2) | | Authors: | Inoue, T, Nishio, N, Kai, Y, Suzuki, S, Kataoka, K. | | Deposit date: | 1999-08-21 | | Release date: | 2000-08-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The significance of the flexible loop in the azurin (Az-iso2) from the obligate methylotroph Methylomonas sp. strain J.
J.Mol.Biol., 333, 2003
|
|
1BQR
 
 | | REDUCED PSEUDOAZURIN | | Descriptor: | COPPER (II) ION, PSEUDOAZURIN | | Authors: | Inoue, T, Nishio, N, Hamanaka, S, Shimomura, T, Harada, S, Suzuki, S, Kohzuma, T, Shidara, S, Iwasaki, H, Kai, Y. | | Deposit date: | 1998-08-17 | | Release date: | 1999-08-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure determinations of oxidized and reduced pseudoazurins from Achromobacter cycloclastes. Concerted movement of copper site in redox forms with the rearrangement of hydrogen bond at a remote histidine.
J.Biol.Chem., 274, 1999
|
|
1BQK
 
 | | OXIDIZED PSEUDOAZURIN | | Descriptor: | COPPER (II) ION, PSEUDOAZURIN | | Authors: | Inoue, T, Nishio, N, Hamanaka, S, Shimomura, T, Harada, S, Suzuki, S, Kohzuma, T, Shidara, S, Iwasaki, H, Kai, Y. | | Deposit date: | 1998-08-17 | | Release date: | 1999-08-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structure determinations of oxidized and reduced pseudoazurins from Achromobacter cycloclastes. Concerted movement of copper site in redox forms with the rearrangement of hydrogen bond at a remote histidine.
J.Biol.Chem., 274, 1999
|
|
4F13
 
 | | Alginate lyase A1-III Y246F complexed with tetrasaccharide | | Descriptor: | 4-deoxy-alpha-L-erythro-hex-4-enopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid, Alginate lyase, beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid | | Authors: | Mikami, B, Ban, M, Suzuki, S, Yoon, H.-J, Miyake, O, Yamasaki, M, Ogura, K, Maruyama, Y, Hashimoto, W, Murata, K. | | Deposit date: | 2012-05-06 | | Release date: | 2012-06-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.208 Å) | | Cite: | Induced-fit motion of a lid loop involved in catalysis in alginate lyase A1-III
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4E1Y
 
 | | Alginate lyase A1-III H192A apo form | | Descriptor: | Alginate lyase | | Authors: | Mikami, B, Ban, M, Suzuki, S, Yoon, H.-J, Miyake, O, Yamasaki, M, Ogura, K, Maruyama, Y, Hashimoto, W, Murata, K. | | Deposit date: | 2012-03-07 | | Release date: | 2012-04-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Induced-fit motion of a lid loop involved in catalysis in alginate lyase A1-III
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4F10
 
 | | Alginate lyase A1-III H192A complexed with tetrasaccharide | | Descriptor: | 4-deoxy-alpha-L-erythro-hex-4-enopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid, Alginate lyase, alpha-L-gulopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid | | Authors: | Mikami, B, Ban, M, Suzuki, S, Yoon, H.-J, Miyake, O, Yamasaki, M, Ogura, K, Maruyama, Y, Hashimoto, W, Murata, K. | | Deposit date: | 2012-05-05 | | Release date: | 2012-06-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Induced-fit motion of a lid loop involved in catalysis in alginate lyase A1-III
Acta Crystallogr.,Sect.D, 68, 2012
|
|
1WG1
 
 | | Solution structure of RNA binding domain in BAB13405(homolog EXC-7) | | Descriptor: | KIAA1579 protein | | Authors: | Tsuda, K, Muto, Y, Nagata, T, Suzuki, S, Someya, T, Kigawa, T, Terada, T, Shirouzu, M, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-27 | | Release date: | 2004-11-27 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Solution structure of RNA binding domain in BAB13405(homolog EXC-7)
To be Published
|
|
1WEL
 
 | | Solution structure of RNA binding domain in NP_006038 | | Descriptor: | RNA-binding protein 12 | | Authors: | Someya, T, Muto, Y, Nagata, T, Suzuki, S, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-25 | | Release date: | 2005-08-23 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of RNA binding domain in NP_006038
To be Published
|
|
2DHT
 
 | |
5H5L
 
 | | Structure of prostaglandin synthase D of Nilaparvata lugens | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLUTATHIONE, ... | | Authors: | Yamamoto, K, Higashiura, A, Suzuki, S, Nakagawa, A. | | Deposit date: | 2016-11-07 | | Release date: | 2017-09-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Molecular structure of a prostaglandin D synthase requiring glutathione from the brown planthopper, Nilaparvata lugens
Biochem. Biophys. Res. Commun., 492, 2017
|
|
3WNM
 
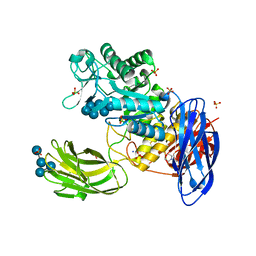 | | D308A mutant of Bacillus circulans T-3040 cycloisomaltooligosaccharide glucanotransferase complexed with isomaltoheptaose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, Cycloisomaltooligosaccharide glucanotransferase, ... | | Authors: | Suzuki, N, Fujimoto, Z, Kim, Y.M, Momma, M, Kishine, N, Suzuki, R, Suzuki, S, Kitamura, S, Kobayashi, M, Kimura, A, Funane, K. | | Deposit date: | 2013-12-10 | | Release date: | 2014-02-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural elucidation of the cyclization mechanism of alpha-1,6-glucan by Bacillus circulans T-3040 cycloisomaltooligosaccharide glucanotransferase.
J.Biol.Chem., 289, 2014
|
|
3WNL
 
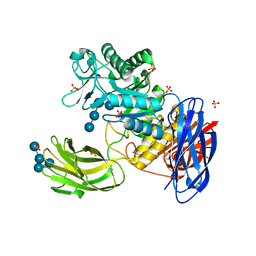 | | D308A mutant of Bacillus circulans T-3040 cycloisomaltooligosaccharide glucanotransferase complexed with isomaltohexaose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, Cycloisomaltooligosaccharide glucanotransferase, ... | | Authors: | Suzuki, N, Fujimoto, Z, Kim, Y.M, Momma, M, Kishine, N, Suzuki, R, Suzuki, S, Kitamura, S, Kobayashi, M, Kimura, A, Funane, K. | | Deposit date: | 2013-12-10 | | Release date: | 2014-02-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural elucidation of the cyclization mechanism of alpha-1,6-glucan by Bacillus circulans T-3040 cycloisomaltooligosaccharide glucanotransferase.
J.Biol.Chem., 289, 2014
|
|
2D0W
 
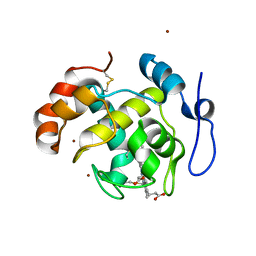 | | Crystal structure of cytochrome cL from Hyphomicrobium denitrificans | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, ZINC ION, cytochrome cL | | Authors: | Nojiri, M, Hira, D, Yamaguchi, K, Suzuki, S. | | Deposit date: | 2005-08-10 | | Release date: | 2006-08-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal structures of cytochrome c(L) and methanol dehydrogenase from Hyphomicrobium denitrificans: structural and mechanistic insights into interactions between the two proteins
Biochemistry, 45, 2006
|
|
2D0V
 
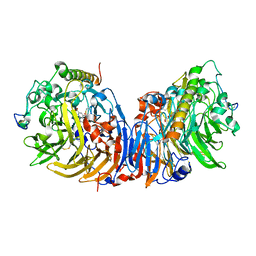 | | Crystal structure of methanol dehydrogenase from Hyphomicrobium denitrificans | | Descriptor: | CALCIUM ION, PYRROLOQUINOLINE QUINONE, methanol dehydrogenase large subunit, ... | | Authors: | Nojiri, M, Hira, D, Yamaguchi, K, Suzuki, S. | | Deposit date: | 2005-08-09 | | Release date: | 2006-08-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Crystal structures of cytochrome c(L) and methanol dehydrogenase from Hyphomicrobium denitrificans: structural and mechanistic insights into interactions between the two proteins
Biochemistry, 45, 2006
|
|
1ZIA
 
 | | OXIDIZED PSEUDOAZURIN | | Descriptor: | COPPER (II) ION, PSEUDOAZURIN | | Authors: | Inoue, T, Nishio, N, Hamanaka, S, Shimomura, T, Harada, S, Suzuki, S, Kohzuma, T, Shidara, S, Iwasaki, H, Kai, Y. | | Deposit date: | 1996-04-09 | | Release date: | 1997-04-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Crystallization and preliminary X-ray studies on pseudoazurin from Achromobacter cycloclastes IAM1013.
J.Biochem.(Tokyo), 114, 1993
|
|
1ZIB
 
 | | REDUCED PSEUDOAZURIN | | Descriptor: | COPPER (II) ION, PSEUDOAZURIN | | Authors: | Inoue, T, Nishio, N, Hamanaka, S, Shimomura, T, Harada, S, Suzuki, S, Kohzuma, T, Shidara, S, Iwasaki, H, Kai, Y. | | Deposit date: | 1996-04-09 | | Release date: | 1997-04-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallization and preliminary X-ray studies on pseudoazurin from Achromobacter cycloclastes IAM1013.
J.Biochem.(Tokyo), 114, 1993
|
|
3WNN
 
 | | D308A mutant of Bacillus circulans T-3040 cycloisomaltooligosaccharide glucanotransferase complexed with isomaltooctaose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, Cycloisomaltooligosaccharide glucanotransferase, ... | | Authors: | Suzuki, N, Fujimoto, Z, Kim, Y.M, Momma, M, Kishine, N, Suzuki, R, Suzuki, S, Kitamura, S, Kobayashi, M, Kimura, A, Funane, K. | | Deposit date: | 2013-12-10 | | Release date: | 2014-02-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural elucidation of the cyclization mechanism of alpha-1,6-glucan by Bacillus circulans T-3040 cycloisomaltooligosaccharide glucanotransferase.
J.Biol.Chem., 289, 2014
|
|
2ROK
 
 | | Solution structure of the cap-binding domain of PARN complexed with the cap analog | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-MONOPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, poly(A)-specific ribonuclease | | Authors: | Nagata, T, Suzuki, S, Endo, R, Shirouzu, M, Terada, T, Inoue, M, Kigawa, T, Guntert, P, Hayashizaki, Y, Muto, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-03-28 | | Release date: | 2009-02-10 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The RRM domain of poly(A)-specific ribonuclease has a noncanonical binding site for mRNA cap analog recognition.
Nucleic Acids Res., 36, 2008
|
|
2DV6
 
 | | Crystal structure of nitrite reductase from Hyphomicrobium denitrificans | | Descriptor: | COPPER (II) ION, Nitrite reductase, POTASSIUM ION | | Authors: | Nojiri, M, Xie, Y, Yamamoto, T, Inoue, T, Suzuki, S, Kai, Y. | | Deposit date: | 2006-07-28 | | Release date: | 2007-02-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and function of a hexameric copper-containing nitrite reductase.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
2DO3
 
 | | Solution structure of the third KOW motif of transcription elongation factor SPT5 | | Descriptor: | Transcription elongation factor SPT5 | | Authors: | Tanabe, W, Suzuki, S, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-04-27 | | Release date: | 2006-10-27 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the third KOW motif of transcription elongation factor SPT5
To be Published
|
|
2DO4
 
 | | Solution structure of the RNA binding domain of squamous cell carcinoma antigen recognized by T cells 3 | | Descriptor: | Squamous cell carcinoma antigen recognized by T-cells 3 | | Authors: | Tanabe, W, Suzuki, S, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-04-27 | | Release date: | 2007-04-17 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the RNA binding domain of squamous cell carcinoma antigen recognized by T cells 3
To be Published
|
|
