4YSA
 
 | | Completely oxidized structure of copper nitrite reductase from Geobacillus thermodenitrificans | | Descriptor: | COPPER (II) ION, Nitrite reductase, SODIUM ION | | Authors: | Fukuda, Y, Tse, K.M, Suzuki, M, Diederichs, K, Hirata, K, Nakane, T, Sugahara, M, Nango, E, Tono, K, Joti, Y, Kameshima, T, Song, C, Hatsui, T, Yabashi, M, Nureki, O, Matsumura, H, Inoue, T, Iwata, S, Mizohata, E. | | Deposit date: | 2015-03-17 | | Release date: | 2016-02-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Redox-coupled structural changes in nitrite reductase revealed by serial femtosecond and microfocus crystallography
J.Biochem., 159, 2016
|
|
4YST
 
 | | Structure of copper nitrite reductase from Geobacillus thermodenitrificans - 24.9 MGy | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, COPPER (II) ION, Nitrite reductase, ... | | Authors: | Fukuda, Y, Tse, K.M, Suzuki, M, Diederichs, K, Hirata, K, Nakane, T, Sugahara, M, Nango, E, Tono, K, Joti, Y, Kameshima, T, Song, C, Hatsui, T, Yabashi, M, Nureki, O, Matsumura, H, Inoue, T, Iwata, S, Mizohata, E. | | Deposit date: | 2015-03-17 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Redox-coupled structural changes in nitrite reductase revealed by serial femtosecond and microfocus crystallography
J.Biochem., 159, 2016
|
|
4YSU
 
 | | Structure of copper nitrite reductase from Geobacillus thermodenitrificans - 25.0 MGy | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, COPPER (II) ION, Nitrite reductase, ... | | Authors: | Fukuda, Y, Tse, K.M, Suzuki, M, Diederichs, K, Hirata, K, Nakane, T, Sugahara, M, Nango, E, Tono, K, Joti, Y, Kameshima, T, Song, C, Hatsui, T, Yabashi, M, Nureki, O, Matsumura, H, Inoue, T, Iwata, S, Mizohata, E. | | Deposit date: | 2015-03-17 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Redox-coupled structural changes in nitrite reductase revealed by serial femtosecond and microfocus crystallography
J.Biochem., 159, 2016
|
|
4YSE
 
 | | High resolution synchrotron structure of copper nitrite reductase from Alcaligenes faecalis | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETIC ACID, COPPER (II) ION, ... | | Authors: | Fukuda, Y, Tse, K.M, Suzuki, M, Diederichs, K, Hirata, K, Nakane, T, Sugahara, M, Nango, E, Tono, K, Joti, Y, Kameshima, T, Song, C, Hatsui, T, Yabashi, M, Nureki, O, Matsumura, H, Inoue, T, Iwata, S, Mizohata, E. | | Deposit date: | 2015-03-17 | | Release date: | 2016-03-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Redox-coupled proton transfer mechanism in nitrite reductase revealed by femtosecond crystallography
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5B35
 
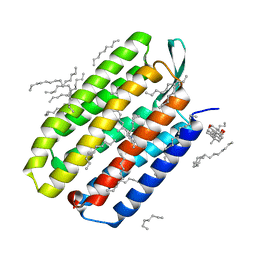 | | Serial Femtosecond Crystallography (SFX) of Ground State Bacteriorhodopsin Crystallized from Bicelles Determined Using 7-keV X-ray Free Electron Laser (XFEL) at SACLA | | Descriptor: | (3R,5S,7R,8R,9S,10S,12S,13R,14S,17R)-10,13-dimethyl-17-[(2R)-pentan-2-yl]-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthrene-3,7,12-triol, Bacteriorhodopsin, DECANE, ... | | Authors: | Mizohata, E, Nakane, T, Suzuki, M. | | Deposit date: | 2016-02-10 | | Release date: | 2016-11-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Membrane protein structure determination by SAD, SIR, or SIRAS phasing in serial femtosecond crystallography using an iododetergent
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
7CE4
 
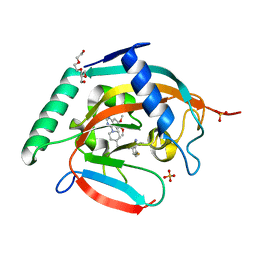 | | Tankyrase2 catalytic domain in complex with K-476 | | Descriptor: | 5-[3-[[1-(6,7-dimethoxyquinazolin-4-yl)piperidin-4-yl]methyl]-2-oxidanylidene-4H-quinazolin-1-yl]-2-fluoranyl-benzenecarbonitrile, Poly [ADP-ribose] polymerase tankyrase-2, SULFATE ION, ... | | Authors: | Takahashi, Y, Suzuki, M, Saito, J. | | Deposit date: | 2020-06-22 | | Release date: | 2021-05-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The dual pocket binding novel tankyrase inhibitor K-476 enhances the efficacy of immune checkpoint inhibitor by attracting CD8 + T cells to tumors.
Am J Cancer Res, 11, 2021
|
|
1SQJ
 
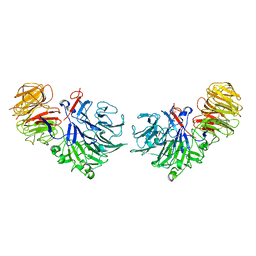 | | Crystal Structure Analysis of Oligoxyloglucan reducing-end-specific cellobiohydrolase (OXG-RCBH) | | Descriptor: | oligoxyloglucan reducing-end-specific cellobiohydrolase | | Authors: | Yaoi, K, Kondo, H, Noro, N, Suzuki, M, Tsuda, S, Mitsuishi, Y. | | Deposit date: | 2004-03-19 | | Release date: | 2004-07-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Tandem Repeat of a Seven-Bladed beta-Propeller Domain in Oligoxyloglucan Reducing-End-Specific Cellobiohydrolase
Structure, 12, 2004
|
|
4YOP
 
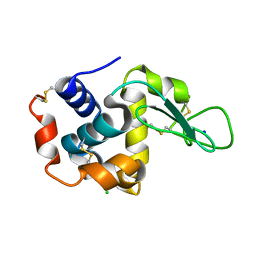 | | CRYSTAL STRUCTURE OF HEN EGG-WHITE LYSOZYME | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Sugahara, M, Nakane, T, Suzuki, M, Nango, E. | | Deposit date: | 2015-03-12 | | Release date: | 2015-12-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Native sulfur/chlorine SAD phasing for serial femtosecond crystallography
Acta Crystallogr.,Sect.D, 71, 2015
|
|
1V5C
 
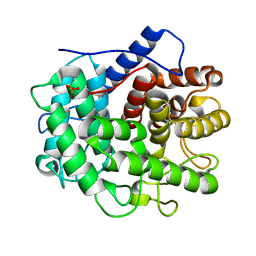 | | The crystal structure of the inactive form chitosanase from Bacillus sp. K17 at pH3.7 | | Descriptor: | SULFATE ION, chitosanase | | Authors: | Adachi, W, Shimizu, S, Sunami, T, Fukazawa, T, Suzuki, M, Yatsunami, R, Nakamura, S, Takenaka, A. | | Deposit date: | 2003-11-22 | | Release date: | 2004-12-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of family GH-8 chitosanase with subclass II specificity from Bacillus sp. K17
J.MOL.BIOL., 343, 2004
|
|
1V5D
 
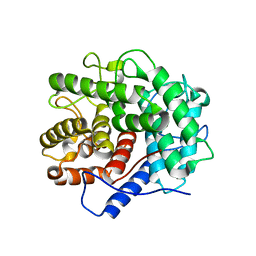 | | The crystal structure of the active form chitosanase from Bacillus sp. K17 at pH6.4 | | Descriptor: | PIPERAZINE-N,N'-BIS(2-ETHANESULFONIC ACID), chitosanase | | Authors: | Adachi, W, Shimizu, S, Sunami, T, Fukazawa, T, Suzuki, M, Yatsunami, R, Nakamura, S, Takenaka, A. | | Deposit date: | 2003-11-22 | | Release date: | 2004-12-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of family GH-8 chitosanase with subclass II specificity from Bacillus sp. K17
J.MOL.BIOL., 343, 2004
|
|
4YM8
 
 | |
5B1G
 
 | |
5B1E
 
 | |
5B1D
 
 | |
5B1F
 
 | |
1RI7
 
 | |
5AZ1
 
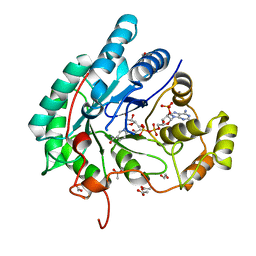 | | Crystal structure of aldo-keto reductase (AKR2E5) complexed with NADPH | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Yamamoto, K, Higashiura, A, Suzuki, M, Nakagawa, A. | | Deposit date: | 2015-09-15 | | Release date: | 2016-02-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural characterization of an aldo-keto reductase (AKR2E5) from the silkworm Bombyx mori
Biochem.Biophys.Res.Commun., 474, 2016
|
|
5AZ0
 
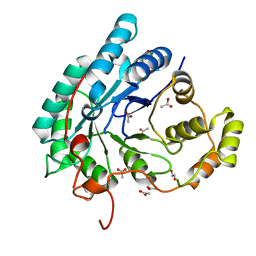 | | Crystal structure of aldo-keto reductase (AKR2E5) of the silkworm, Bombyx mori | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Yamamoto, K, Higashiura, A, Suzuki, M, Nakagawa, A. | | Deposit date: | 2015-09-15 | | Release date: | 2016-02-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural characterization of an aldo-keto reductase (AKR2E5) from the silkworm Bombyx mori
Biochem.Biophys.Res.Commun., 474, 2016
|
|
5B22
 
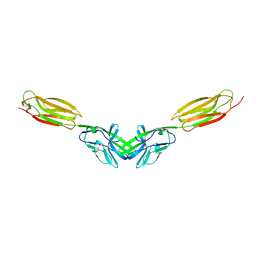 | | Dimer structure of murine Nectin-3 D1D2 | | Descriptor: | Nectin-3, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Takebe, K, Sangawa, T, Katsutani, T, Narita, H, Suzuki, M. | | Deposit date: | 2015-12-28 | | Release date: | 2016-12-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Dimer structure of murine Nectin-3 D1D2
To Be Published
|
|
5B21
 
 | |
6L9C
 
 | | Neutron structure of copper amine oxidase from Arthrobacter glibiformis at pD 7.4 | | Descriptor: | COPPER (II) ION, Phenylethylamine oxidase, SODIUM ION | | Authors: | Murakawa, T, Kurihara, K, Shoji, M, Shibazaki, C, Sunami, T, Tamada, T, Yano, N, Yamada, T, Kusaka, K, Suzuki, M, Shigeta, Y, Kuroki, R, Hayashi, H, Yano, Y, Tanizawa, K, Adachi, M, Okajima, T. | | Deposit date: | 2019-11-08 | | Release date: | 2020-04-29 | | Last modified: | 2023-11-22 | | Method: | NEUTRON DIFFRACTION (1.14 Å), X-RAY DIFFRACTION | | Cite: | Neutron crystallography of copper amine oxidase reveals keto/enolate interconversion of the quinone cofactor and unusual proton sharing.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7XY8
 
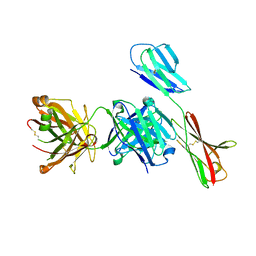 | | Crystal structure of antibody Fab fragment in complex with CD147(EMMPIRIN) | | Descriptor: | Isoform 2 of Basigin, heavy chain, light chain | | Authors: | Nakamura, K, Amano, M, Yoneda, K, Suzuki, M, Fukuchi, K. | | Deposit date: | 2022-06-01 | | Release date: | 2022-11-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Novel Antibody Exerts Antitumor Effect through Downregulation of CD147 and Activation of Multiple Stress Signals.
J Oncol, 2022, 2022
|
|
5Y1A
 
 | | HBP35 of Porphyromonas gingivalis | | Descriptor: | 35 kDa hemin binding protein | | Authors: | Kakuda, S, Suzuki, M, Sato, K. | | Deposit date: | 2017-07-20 | | Release date: | 2018-07-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Immunoglobulin-like domains of the cargo proteins are essential for protein stability during secretion by the type IX secretion system.
Mol. Microbiol., 110, 2018
|
|
5YYP
 
 | | Structure K137A thaumatin | | Descriptor: | GLYCEROL, L(+)-TARTARIC ACID, Preprothaumatin I | | Authors: | Masuda, T, Kigo, S, Mitsumoto, M, Ohta, K, Suzuki, M, Mikami, B, Kitabatake, N, Tani, F. | | Deposit date: | 2017-12-10 | | Release date: | 2018-03-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.01 Å) | | Cite: | Positive Charges on the Surface of Thaumatin Are Crucial for the Multi-Point Interaction with the Sweet Receptor.
Front Mol Biosci, 5, 2018
|
|
5YYQ
 
 | | Structure K78A thaumatin | | Descriptor: | GLYCEROL, L(+)-TARTARIC ACID, Preprothaumatin I | | Authors: | Masuda, T, Kigo, S, Mitsumoto, M, Ohta, K, Suzuki, M, Mikami, B, Kitabatake, N, Tani, F. | | Deposit date: | 2017-12-10 | | Release date: | 2018-03-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Positive Charges on the Surface of Thaumatin Are Crucial for the Multi-Point Interaction with the Sweet Receptor.
Front Mol Biosci, 5, 2018
|
|
