4P6A
 
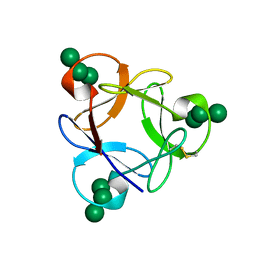 | | Crystal structure of a potent anti-HIV lectin actinohivin in complex with alpha-1,2-mannotriose | | Descriptor: | Actinohivin, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose | | Authors: | Zhang, F, Hoque, M.M, Suzuki, K, Tsunoda, M, Naomi, O, Tanaka, H, Takenaka, A. | | Deposit date: | 2014-03-23 | | Release date: | 2015-03-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.398 Å) | | Cite: | The characteristic structure of anti-HIV actinohivin in complex with three HMTG D1 chains of HIV-gp120.
Chembiochem, 15, 2014
|
|
7DQC
 
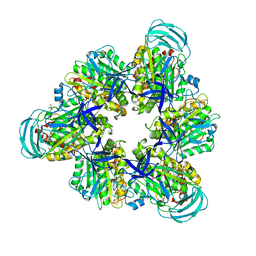 | | Crystal structure of nucleotide-free mutant A(S23C)3B(N64C)3 complex from Enterococcus hirae V-ATPase | | Descriptor: | GLYCEROL, V-type sodium ATPase catalytic subunit A, V-type sodium ATPase subunit B | | Authors: | Maruyama, S, Suzuki, K, Mizutani, K, Imai, F.L, Ishizuka-Katsura, Y, Shirouzu, M, Murata, M. | | Deposit date: | 2020-12-23 | | Release date: | 2021-12-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.706 Å) | | Cite: | The combination of high-speed AFM and X-ray crystallography reveals rotary catalytic mechanism of shaftless V1-ATPase
To Be Published
|
|
7DQE
 
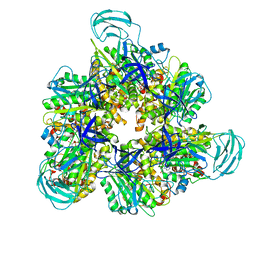 | | Crystal structure of the ADP-bound mutant A(S23C)3B(N64C)3 complex from enterococcus hirae V-ATPase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Maruyama, S, Nakamoto, K, Suzuki, K, Mizutani, K, Imai, F.L, Ishizuka-Katsura, Y, Shirouzu, M, Murata, T. | | Deposit date: | 2020-12-23 | | Release date: | 2021-12-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.694 Å) | | Cite: | The Combination of High-Speed AFM and X-ray Crystallography Reveals Rotary Catalytic Mechanism of Shaftless V1-ATPase
To Be Published
|
|
7CK5
 
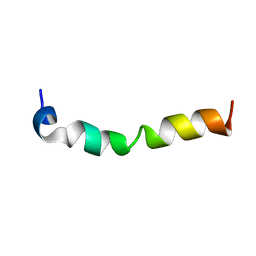 | | Solution structure of 28 amino acid polypeptide (354-381) in Plantago asiatica mosaic virus replicase bound to SDS micelle | | Descriptor: | PlAMV replicase peptide from RNA-dependent RNA polymerase | | Authors: | Komatsu, K, Sasaki, N, Yoshida, T, Suzuki, K, Masujima, Y, Hashimoto, M, Watanabe, S, Tochio, N, Kigawa, T, Yamaji, Y, Oshima, K, Namba, S, Nelson, R, Arie, T. | | Deposit date: | 2020-07-15 | | Release date: | 2021-07-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Identification of a Proline-Kinked Amphipathic alpha-Helix Downstream from the Methyltransferase Domain of a Potexvirus Replicase and Its Role in Virus Replication and Perinuclear Complex Formation.
J.Virol., 95, 2021
|
|
4IJ0
 
 | | Structures of DNA duplexes containing O6-carboxymethylguanine, a lesion associated with gastrointestinal cancer, reveal a mechanism for inducing transition mutation | | Descriptor: | 2'-(4-HYDROXYPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, DNA (5'-D(*CP*GP*CP*(C6G)P*AP*AP*TP*TP*CP*GP*CP*G)-3'), STRONTIUM ION | | Authors: | Zhang, F, Suzuki, K, Tsunoda, M, Wilkinson, O, Millington, C.L, Williams, D.M, Morishita, E.C, Takenaka, A. | | Deposit date: | 2012-12-20 | | Release date: | 2013-05-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structures of DNA duplexes containing O6-carboxymethylguanine, a lesion associated with gastrointestinal cancer, reveal a mechanism for inducing pyrimidine transition mutations
Nucleic Acids Res., 41, 2013
|
|
4ITD
 
 | | Structures of DNA duplexes containing O6-carboxymethylguanine, a lesion associated with gastrointestinal cancer, reveal a mechanism for inducing transition mutation | | Descriptor: | 2'-(4-HYDROXYPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, DNA (5'-D(*CP*GP*CP*GP*(C6G)P*AP*TP*TP*CP*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Zhang, F, Suzuki, K, Tsunoda, M, Wilkinson, O, Millington, C.L, Williams, D.M, Morishita, E.C, Takenaka, A. | | Deposit date: | 2013-01-18 | | Release date: | 2013-05-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structures of DNA duplexes containing O6-carboxymethylguanine, a lesion associated with gastrointestinal cancer, reveal a mechanism for inducing pyrimidine transition mutations
Nucleic Acids Res., 41, 2013
|
|
7X5F
 
 | | Nrf2-MafG heterodimer bound with CsMBE2 | | Descriptor: | Nuclear factor erythroid 2-related factor 2, Synthetic DNA, Transcription factor MafG | | Authors: | Sengoku, T, Shiina, M, Suzuki, K, Hamada, K, Sato, K, Uchiyama, A, Okada, C, Baba, S, Ohta, T, Motohashi, H, Yamamoto, M, Ogata, K. | | Deposit date: | 2022-03-04 | | Release date: | 2022-11-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of transcription regulation by CNC family transcription factor, Nrf2.
Nucleic Acids Res., 50, 2022
|
|
7X5E
 
 | | Nrf2-MafG heterodimer bound with CsMBE1 | | Descriptor: | DNA (5'-D(*CP*AP*TP*GP*AP*TP*GP*AP*GP*TP*CP*AP*GP*CP*AP*A)-3'), DNA (5'-D(*GP*TP*TP*GP*CP*TP*GP*AP*CP*TP*CP*AP*TP*CP*AP*T)-3'), HEXAETHYLENE GLYCOL, ... | | Authors: | Sengoku, T, Shiina, M, Suzuki, K, Hamada, K, Sato, K, Uchiyama, A, Okada, C, Baba, S, Ohta, T, Motohashi, H, Yamamoto, M, Ogata, K. | | Deposit date: | 2022-03-04 | | Release date: | 2022-11-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of transcription regulation by CNC family transcription factor, Nrf2.
Nucleic Acids Res., 50, 2022
|
|
7X5G
 
 | | Nrf2 (A510Y)-MafG heterodimer bound with CsMBE2 | | Descriptor: | DNA (5'-D(*CP*AP*CP*AP*GP*TP*GP*AP*CP*TP*CP*AP*GP*CP*AP*G)-3'), DNA (5'-D(*GP*CP*TP*GP*CP*TP*GP*AP*GP*TP*CP*AP*CP*TP*GP*T)-3'), Nuclear factor erythroid 2-related factor 2, ... | | Authors: | Sengoku, T, Shiina, M, Suzuki, K, Hamada, K, Sato, K, Uchiyama, A, Okada, C, Baba, S, Ohta, T, Motohashi, H, Yamamoto, M, Ogata, K. | | Deposit date: | 2022-03-04 | | Release date: | 2022-11-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of transcription regulation by CNC family transcription factor, Nrf2.
Nucleic Acids Res., 50, 2022
|
|
6M4B
 
 | |
1V28
 
 | | Solution structure of paralytic peptide of the wild Silkmoth, Antheraea yamamai | | Descriptor: | Paralytic Peptide | | Authors: | Kawaguchi, K, Ying, A, Suzuki, K, Kumaki, Y, Demura, M, Nitta, K. | | Deposit date: | 2003-10-09 | | Release date: | 2004-10-26 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of paralytic peptide of the wild Silkmoth Antheraea yamamai by NMR
To be Published
|
|
6IIE
 
 | | Crystal structure of human diacylglycerol kinase alpha EF-hand domains bound to Ca2+ | | Descriptor: | CALCIUM ION, Diacylglycerol kinase alpha, GLYCEROL, ... | | Authors: | Takahashi, D, Suzuki, K, Sakamoto, T, Iwamoto, T, Murata, T, Sakane, F. | | Deposit date: | 2018-10-04 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.142 Å) | | Cite: | Crystal structure and calcium-induced conformational changes of diacylglycerol kinase alpha EF-hand domains.
Protein Sci., 28, 2019
|
|
1V5B
 
 | | The Structure Of The Mutant, S225A and E251L, Of 3-Isopropylmalate Dehydrogenase From Bacillus Coagulans | | Descriptor: | 3-isopropylmalate dehydrogenase, SULFATE ION | | Authors: | Fujita, K, Minami, H, Suzuki, K, Tsunoda, M, Sekiguchi, T, Mizui, R, Tsuzaki, S, Nakamura, S, Takenaka, A. | | Deposit date: | 2003-11-22 | | Release date: | 2005-02-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal structure of a highly thermo-stabilized mutant of 3-isopropylmalate dehydrogenase from Bacillus coagulans: An evaluation of local packing density in the hydrophobic core
To be Published
|
|
1JEH
 
 | | CRYSTAL STRUCTURE OF YEAST E3, LIPOAMIDE DEHYDROGENASE | | Descriptor: | DIHYDROLIPOAMIDE DEHYDROGENASE, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Toyoda, T, Suzuki, K, Sekigushi, T, Reed, J, Takenaka, A. | | Deposit date: | 2001-06-18 | | Release date: | 2001-07-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of eucaryotic E3, lipoamide dehydrogenase from yeast.
J.Biochem., 123, 1998
|
|
5ZEA
 
 | | Crystal structure of the nucleotide-free mutant A3B3 | | Descriptor: | GLYCEROL, V-type sodium ATPase catalytic subunit A, V-type sodium ATPase subunit B | | Authors: | Maruyama, S, Suzuki, K, Mizutani, K, Saito, Y, Imai, F.L, Ishizuka-Katsura, Y, Shirouzu, M, Ichiro, Y, Murata, T. | | Deposit date: | 2018-02-27 | | Release date: | 2019-02-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.384 Å) | | Cite: | Metastable asymmetrical structure of a shaftless V1motor.
Sci Adv, 5, 2019
|
|
1V53
 
 | | The crystal structure of 3-isopropylmalate dehydrogenase from Bacillus coagulans | | Descriptor: | 3-isopropylmalate dehydrogenase | | Authors: | Fujita, K, Minami, H, Suzuki, K, Tsunoda, M, Sekiguchi, T, Mizui, R, Tsuzaki, S, Nakamura, S, Takenaka, A. | | Deposit date: | 2003-11-20 | | Release date: | 2005-02-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | The crystal structure of 3-isopropylmalate dehydrogenase from Bacillus coagulans
To be Published
|
|
1VAX
 
 | | Crystal Structure of Uricase from Arthrobacter globiformis | | Descriptor: | Uric acid oxidase | | Authors: | Hossain, M.T, Suzuki, K, Yamamoto, T, Imamura, S, Sekiguchi, T, Takenaka, A. | | Deposit date: | 2004-02-19 | | Release date: | 2005-06-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystal Structure of Uricase from Arthrobacter globiformis
To be Published
|
|
1VAY
 
 | | Crystal Structure of Uricase from Arthrobacter globiformis with inhibitor 8-azaxanthine | | Descriptor: | 8-AZAXANTHINE, Uric acid oxidase | | Authors: | Hossain, M.T, Suzuki, K, Yamamoto, T, Imamura, S, Sekiguchi, T, Takenaka, A. | | Deposit date: | 2004-02-19 | | Release date: | 2005-06-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Crystal Structure of Uricase from Arthrobacter Globiformis Complexed with an inhibitor 8-Azaxanthine
To be Published
|
|
5XA3
 
 | | Crystal Structure of P450BM3 with Benzyloxycarbonyl-L-prolyl-L-phenylalanine | | Descriptor: | Bifunctional cytochrome P450/NADPH-P450 reductase, DIMETHYL SULFOXIDE, PHENYLALANINE, ... | | Authors: | Shoji, O, Yanagisawa, S, Stanfield, J.K, Suzuki, K, Kasai, C, Cong, Z, Sugimoto, H, Shiro, Y, Watanabe, Y. | | Deposit date: | 2017-03-10 | | Release date: | 2018-02-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Direct Hydroxylation of Benzene to Phenol by Cytochrome P450BM3 Triggered by Amino Acid Derivatives.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5ZE9
 
 | | Crystal structure of AMP-PNP bound mutant A3B3 complex from Enterococcus hirae V-ATPase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Maruyama, S, Suzuki, K, Sasaki, H, Mizutani, K, Saito, Y, Imai, F.L, Ishizuka-Katsura, Y, Shirouzu, M, Ichiro, Y, Murata, T. | | Deposit date: | 2018-02-27 | | Release date: | 2019-02-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Metastable asymmetrical structure of a shaftless V1motor.
Sci Adv, 5, 2019
|
|
1WYE
 
 | | Crystal structure of 2-keto-3-deoxygluconate kinase (form 1) from Sulfolobus Tokodaii | | Descriptor: | 2-keto-3-deoxygluconate kinase | | Authors: | Toyoda, T, Suzuki, K, Hossain, M.T, Koike, I, Sekiguchi, T, Takenaka, A. | | Deposit date: | 2005-02-12 | | Release date: | 2006-07-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of 2-keto-3-deoxygluconate kinase from Sulfolobus Tokodaii
To be Published
|
|
2DTM
 
 | | Thermodynamic and structural analyses of hydrolytic mechanism by catalytic antibodies | | Descriptor: | IMMUNOGLOBULIN 6D9 | | Authors: | Oda, M, Ito, N, Tsumuraya, T, Suzuki, K, Fujii, I. | | Deposit date: | 2006-07-13 | | Release date: | 2007-05-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Thermodynamic and structural basis for transition-state stabilization in antibody-catalyzed hydrolysis
J.Mol.Biol., 369, 2007
|
|
1V59
 
 | | Crystal structure of yeast lipoamide dehydrogenase complexed with NAD+ | | Descriptor: | Dihydrolipoamide dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Adachi, W, Suzuki, K, Tsunoda, M, Sekiguchi, T, Reed, L.J, Takenaka, A. | | Deposit date: | 2003-11-21 | | Release date: | 2005-02-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of yeast lipoamide dehydrogenase complexed with NAD+
To be Published
|
|
1V5G
 
 | | Crystal Structure of the Reaction Intermediate between Pyruvate oxidase containing FAD and TPP, and Substrate Pyruvate | | Descriptor: | 2-ACETYL-THIAMINE DIPHOSPHATE, FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION, ... | | Authors: | Hossain, M.T, Suzuki, K, Yamamoto, T, Imamura, S, Sekiguchi, T, Takenaka, A. | | Deposit date: | 2003-11-22 | | Release date: | 2005-06-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | The structures of pyruvate oxidase from Aerococcus viridans with cofactors and with a reaction intermediate reveal the flexibility of the active-site tunnel for catalysis.
Acta Crystallogr.,Sect.F, 63, 2007
|
|
1V5F
 
 | | Crystal Structure of Pyruvate oxidase complexed with FAD and TPP, from Aerococcus viridans | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION, Pyruvate oxidase, ... | | Authors: | Hossain, M.T, Suzuki, K, Yamamoto, T, Imamura, S, Sekiguchi, T, Takenaka, A. | | Deposit date: | 2003-11-22 | | Release date: | 2005-06-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structures of pyruvate oxidase from Aerococcus viridans with cofactors and with a reaction intermediate reveal the flexibility of the active-site tunnel for catalysis.
Acta Crystallogr.,Sect.F, 63, 2007
|
|
