1UCG
 
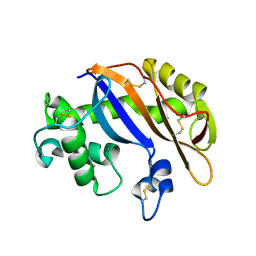 | | Crystal structure of Ribonuclease MC1 N71T mutant | | Descriptor: | MANGANESE (II) ION, Ribonuclease MC | | Authors: | Suzuki, A, Numata, T, Yao, M, Tanaka, I, Kimura, M. | | Deposit date: | 2003-04-14 | | Release date: | 2003-04-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structures of the ribonuclease MC1 mutants N71T and N71S in complex with 5'-GMP: structural basis for alterations in substrate specificity
Biochemistry, 42, 2003
|
|
1UCD
 
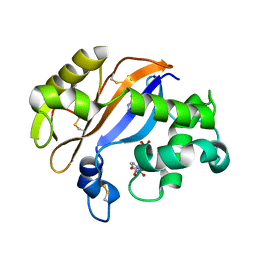 | | Crystal structure of Ribonuclease MC1 from bitter gourd seeds complexed with 5'-UMP | | Descriptor: | Ribonuclease MC, URACIL, URIDINE-5'-MONOPHOSPHATE | | Authors: | Suzuki, A, Numata, T, Yao, M, Kimura, M, Tanaka, I. | | Deposit date: | 2003-04-10 | | Release date: | 2004-05-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure of RNase MC1 from bitter gourd seeds in complex with 5'UMP
To be published
|
|
1UCA
 
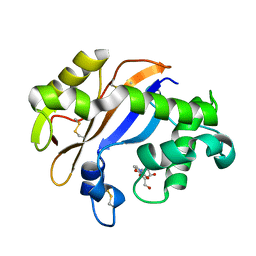 | | Crystal structure of the Ribonuclease MC1 from bitter gourd seeds complexed with 2'-UMP | | Descriptor: | PHOSPHORIC ACID MONO-[2-(2,4-DIOXO-3,4-DIHYDRO-2H-PYRIMIDIN-1-YL)-4-HYDROXY-5-HYDROXYMETHYL-TETRAHYDRO-FURAN-3-YL] ESTER, Ribonuclease MC | | Authors: | Suzuki, A, Yao, M, Tanaka, I, Numata, T, Kikukawa, S, Yamasaki, N, Kimura, M. | | Deposit date: | 2003-04-10 | | Release date: | 2003-04-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystal structures of the ribonuclease MC1 from bitter gourd seeds, complexed with 2'-UMP or 3'-UMP, reveal structural basis for uridine specificity
Biochem.Biophys.Res.Commun., 275, 2000
|
|
1UCC
 
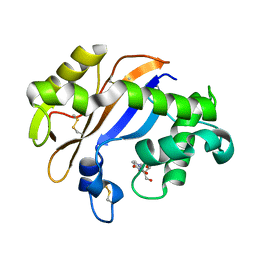 | | Crystal structure of the Ribonuclease MC1 from bitter gourd seeds complexed with 3'-UMP. | | Descriptor: | 3'-URIDINEMONOPHOSPHATE, Ribonuclease MC | | Authors: | Suzuki, A, Yao, M, Tanaka, I, Numata, T, Kikukawa, S, Yamasaki, N, Kimura, M. | | Deposit date: | 2003-04-10 | | Release date: | 2003-04-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structures of the ribonuclease MC1 from bitter gourd seeds, complexed with 2'-UMP or 3'-UMP, reveal structural basis for uridine specificity
Biochem.Biophys.Res.Commun., 275, 2000
|
|
2ZE4
 
 | | Crystal structure of phospholipase D from streptomyces antibioticus | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Phospholipase D | | Authors: | Suzuki, A, Kakuno, K, Saito, R, Iwasaki, Y, Yamane, T, Yamane, T. | | Deposit date: | 2007-12-05 | | Release date: | 2007-12-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of phospholipase D from streptomyces antibioticus
To be Published
|
|
2ZE9
 
 | | Crystal structure of H168A mutant of phospholipase D from Streptomyces antibioticus, as a complex with phosphatidylcholine | | Descriptor: | (2R)-3-(phosphonooxy)propane-1,2-diyl diheptanoate, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Phospholipase D | | Authors: | Suzuki, A, Toda, H, Iwasaki, Y, Yamane, T, Yamane, T. | | Deposit date: | 2007-12-06 | | Release date: | 2007-12-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of phospholipase D from streptomyces antibioticus
To be Published
|
|
3B0C
 
 | | Crystal structure of the chicken CENP-T histone fold/CENP-W complex, crystal form I | | Descriptor: | CITRIC ACID, Centromere protein T, Centromere protein W | | Authors: | Nishino, T, Takeuchi, K, Gascoigne, K.E, Suzuki, A, Hori, T, Oyama, T, Morikawa, K, Cheeseman, I.M, Fukagawa, T. | | Deposit date: | 2011-06-08 | | Release date: | 2012-03-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | CENP-T-W-S-X Forms a Unique Centromeric Chromatin Structure with a Histone-like Fold.
Cell(Cambridge,Mass.), 148, 2012
|
|
3B0B
 
 | | Crystal structure of the chicken CENP-S/CENP-X complex | | Descriptor: | Centromere protein S, Centromere protein X | | Authors: | Nishino, T, Takeuchi, K, Gascoigne, K.E, Suzuki, A, Hori, T, Oyama, T, Morikawa, K, Cheeseman, I.M, Fukagawa, T. | | Deposit date: | 2011-06-08 | | Release date: | 2012-03-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | CENP-T-W-S-X Forms a Unique Centromeric Chromatin Structure with a Histone-like Fold.
Cell(Cambridge,Mass.), 148, 2012
|
|
1VDQ
 
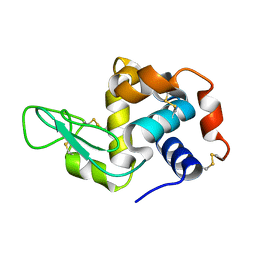 | | The crystal structure of the orthorhombic form of hen egg white lysozyme at 1.5 angstroms resolution | | Descriptor: | Lysozyme C | | Authors: | Aibara, S, Suzuki, A, Kidera, A, Shibata, K, Yamane, T, DeLucas, L.J, Hirose, M. | | Deposit date: | 2004-03-24 | | Release date: | 2004-04-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structure of the orthorhombic form of hen egg white lysozyme at 1.5 angstroms resolution
to be published
|
|
1VDT
 
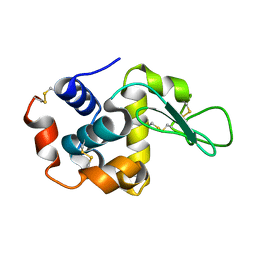 | | The crystal structure of the tetragonal form of hen egg white lysozyme at 1.7 angstroms resolution under basic conditions in space | | Descriptor: | Lysozyme C | | Authors: | Aibara, S, Suzuki, A, Kidera, A, Shibata, K, Yamane, T, DeLucas, L.J, Hirose, M. | | Deposit date: | 2004-03-24 | | Release date: | 2004-04-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structure of the tetragonal form of hen egg white lysozyme at 1.7 angstroms resolution under basic conditions in space
to be published
|
|
1VED
 
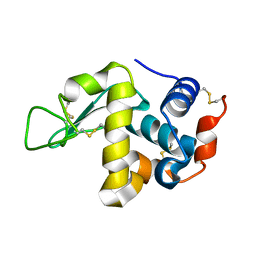 | | The crystal structure of the orthorhombic form of hen egg white lysozyme at 1.9 angstroms resolution in space | | Descriptor: | Lysozyme C | | Authors: | Aibara, S, Suzuki, A, Kidera, A, Shibata, K, Yamane, T, DeLucas, L.J, Hirose, M. | | Deposit date: | 2004-03-30 | | Release date: | 2004-04-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of the orthorhombic form of hen egg white lysozyme at 1.9 angstroms resolution in space
To be Published
|
|
1VDS
 
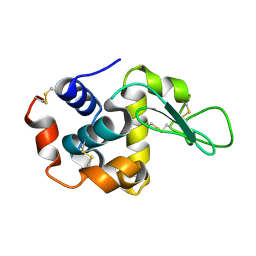 | | The crystal structure of the tetragonal form of hen egg white lysozyme at 1.6 angstroms resolution in space | | Descriptor: | Lysozyme C | | Authors: | Aibara, S, Suzuki, A, Kidera, A, Shibata, K, Yamane, T, DeLucas, L.J, Hirose, M. | | Deposit date: | 2004-03-24 | | Release date: | 2004-04-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of the tetragonal form of hen egg white lysozyme at 1.6 angstroms resolution in space
to be published
|
|
5B18
 
 | | Crystal Structure of a Darunavir Resistant HIV-1 Protease | | Descriptor: | ACETATE ION, CHLORIDE ION, Protease | | Authors: | Suzuki, K, Ode, H, Nakashima, M, Sugiura, W, Watanabe, N, Suzuki, A, Iwatani, Y. | | Deposit date: | 2015-11-30 | | Release date: | 2016-04-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Unique Flap Conformation in an HIV-1 Protease with High-Level Darunavir Resistance
Front Microbiol, 7, 2016
|
|
3HJE
 
 | | Crystal structure of sulfolobus tokodaii hypothetical maltooligosyl trehalose synthase | | Descriptor: | 704aa long hypothetical glycosyltransferase, GLYCEROL | | Authors: | Cielo, C.B.C, Okazaki, S, Suzuki, A, Mizushima, T, Masui, R, Kuramitsu, S, Yamane, T. | | Deposit date: | 2009-05-21 | | Release date: | 2010-04-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of ST0929, a putative glycosyl transferase from Sulfolobus tokodaii
Acta Crystallogr.,Sect.F, 66, 2010
|
|
1J1G
 
 | | Crystal structure of the RNase MC1 mutant N71S in complex with 5'-GMP | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, Ribonuclease MC1 | | Authors: | Numata, T, Suzuki, A, Kakuta, Y, Kimura, K, Yao, M, Tanaka, I, Yoshida, Y, Ueda, T, Kimura, M. | | Deposit date: | 2002-12-04 | | Release date: | 2003-05-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structures of the Ribonuclease MC1 Mutants N71T and N71S in Complex with 5'-GMP: Structural Basis for Alterations in Substrate Specificity
Biochemistry, 42, 2003
|
|
1J1F
 
 | | Crystal structure of the RNase MC1 mutant N71T in complex with 5'-GMP | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, RIBONUCLEASE MC1 | | Authors: | Numata, T, Suzuki, A, Kakuta, Y, Kimura, K, Yao, M, Tanaka, I, Yoshida, Y, Ueda, T, Kimura, M. | | Deposit date: | 2002-12-03 | | Release date: | 2003-05-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structures of the Ribonuclease MC1 Mutants N71T and N71S in Complex with 5'-GMP: Structural Basis for Alterations in Substrate Specificity
Biochemistry, 42, 2003
|
|
3DXW
 
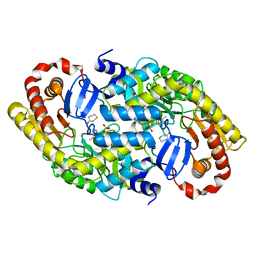 | | The crystal structure of alpha-amino-epsilon-caprolactam racemase from Achromobacter obae complexed with epsilon caprolactam | | Descriptor: | Alpha-amino-epsilon-caprolactam racemase, PYRIDOXAL-5'-PHOSPHATE, azepan-2-one | | Authors: | Okazaki, S, Suzuki, A, Komeda, H, Asano, Y, Yamane, T. | | Deposit date: | 2008-07-25 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | The novel structure of a pyridoxal 5'-phosphate-dependent fold-type I racemase, alpha-amino-epsilon-caprolactam racemase from Achromobacter obae
Biochemistry, 48, 2009
|
|
3DXV
 
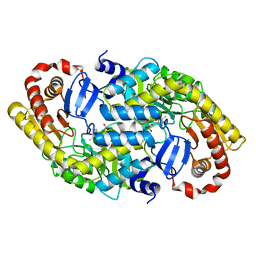 | | The crystal structure of alpha-amino-epsilon-caprolactam racemase from Achromobacter obae | | Descriptor: | Alpha-amino-epsilon-caprolactam racemase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Okazaki, S, Suzuki, A, Komeda, H, Asano, Y, Yamane, T. | | Deposit date: | 2008-07-25 | | Release date: | 2009-02-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | The novel structure of a pyridoxal 5'-phosphate-dependent fold-type I racemase, alpha-amino-epsilon-caprolactam racemase from Achromobacter obae
Biochemistry, 48, 2009
|
|
8WF5
 
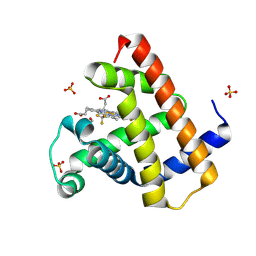 | | Horse heart myoglobin reconstituted with an iron complex of porphyrin bearing two CF3 groups (rMb(FePor(CF3)2)) | | Descriptor: | GLYCEROL, Myoglobin, PROTOPORPHYRIN(CF3)2 CONTAINING FE, ... | | Authors: | Kagawa, Y, Oohora, K, Himiyama, T, Suzuki, A, Hayashi, T. | | Deposit date: | 2023-09-19 | | Release date: | 2024-06-12 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Redox Engineering of Myoglobin by Cofactor Substitution to Enhance Cyclopropanation Reactivity.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
1WMX
 
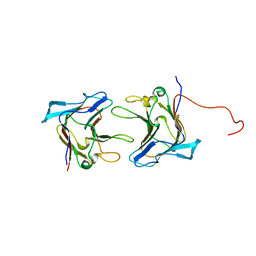 | | Crystal Structure of Family 30 Carbohydrate Binding Module | | Descriptor: | COG3291: FOG: PKD repeat, SULFATE ION | | Authors: | Horiguchi, Y, Kono, M, Suzuki, A, Yamane, T, Arai, M, Sakka, K, Omiya, K. | | Deposit date: | 2004-07-21 | | Release date: | 2004-08-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Family 30 Carbohydrate Binding Module
To be Published
|
|
1WSD
 
 | | Alkaline M-protease form I crystal structure | | Descriptor: | CALCIUM ION, M-protease, SULFATE ION | | Authors: | Shirai, T, Suzuki, A, Yamane, T, Ashida, T, Kobayashi, T, Hitomi, J, Ito, S. | | Deposit date: | 2004-11-05 | | Release date: | 2004-11-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | High-resolution crystal structure of M-protease: phylogeny aided analysis of the high-alkaline adaptation mechanism
Protein Eng., 10, 1997
|
|
3P5V
 
 | | Actinidin from Actinidia arguta planch (Sarusashi) | | Descriptor: | Actinidin, CADMIUM ION | | Authors: | Manickam, Y, Nirmal, N, Suzuki, A, Sugiyama, Y, Yamane, T, Devadasan, V, Sharma, A. | | Deposit date: | 2010-10-11 | | Release date: | 2010-11-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis of actinidin and a comparison of cadmium and sulfur anomalous signals from actinidin crystals measured using in-house copper- and chromium-anode X-ray sources
Acta Crystallogr.,Sect.D, 66, 2010
|
|
1WZX
 
 | | Crystal Structure of Family 30 Carbohydrate Binding Module. | | Descriptor: | COG3291: FOG: PKD repeat | | Authors: | Horiguchi, Y, Kono, M, Suzuki, A, Yamane, T, Arai, M, Sakka, K, Omiya, K. | | Deposit date: | 2005-03-10 | | Release date: | 2005-03-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.52 Å) | | Cite: | Crystal Structure of Family 30 Carbohydrate Binding Module
To be Published
|
|
1MPT
 
 | | CRYSTAL STRUCTURE OF A NEW ALKALINE SERINE PROTEASE (M-PROTEASE) FROM BACILLUS SP. KSM-K16 | | Descriptor: | CALCIUM ION, M-PROTEASE | | Authors: | Yamane, T, Kani, T, Hatanaka, T, Suzuki, A, Ashida, T, Kobayashi, T, Ito, S, Yamashita, O. | | Deposit date: | 1994-04-13 | | Release date: | 1994-06-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of a new alkaline serine protease (M-protease) from Bacillus sp. KSM-K16.
Acta Crystallogr.,Sect.D, 51, 1995
|
|
3P5W
 
 | | Actinidin from Actinidia arguta planch (Sarusashi) | | Descriptor: | Actinidin, CADMIUM ION | | Authors: | Manickam, Y, Nirmal, N, Suzuki, A, Sugiyama, Y, Yamane, T, Devadasan, V, Sharma, A. | | Deposit date: | 2010-10-11 | | Release date: | 2010-11-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural analysis of actinidin and a comparison of cadmium and sulfur anomalous signals from actinidin crystals measured using in-house copper- and chromium-anode X-ray sources
Acta Crystallogr.,Sect.D, 66, 2010
|
|
