5HBN
 
 | |
6FH1
 
 | | Protein arginine kinase McsB in the apo state | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, IMIDAZOLE, ... | | Authors: | Suskiewicz, M.J, Heuck, A, Vu, L.D, Clausen, T. | | Deposit date: | 2018-01-12 | | Release date: | 2019-02-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of McsB, a protein kinase for regulated arginine phosphorylation.
Nat.Chem.Biol., 15, 2019
|
|
6FH4
 
 | |
6FH2
 
 | | Protein arginine kinase McsB in the AMP-PN-bound state | | Descriptor: | 1,2-ETHANEDIOL, AMP PHOSPHORAMIDATE, Protein-arginine kinase | | Authors: | Suskiewicz, M.J, Heuck, A, Vu, L.D, Clausen, T. | | Deposit date: | 2018-01-12 | | Release date: | 2019-02-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of McsB, a protein kinase for regulated arginine phosphorylation.
Nat.Chem.Biol., 15, 2019
|
|
6FH3
 
 | | Protein arginine kinase McsB in the pArg-bound state | | Descriptor: | 1,2-ETHANEDIOL, Protein-arginine kinase, phospho-arginine | | Authors: | Suskiewicz, M.J, Heuck, A, Vu, L.D, Clausen, T. | | Deposit date: | 2018-01-12 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of McsB, a protein kinase for regulated arginine phosphorylation.
Nat.Chem.Biol., 15, 2019
|
|
6TX1
 
 | | HPF1 from Nematostella vectensis | | Descriptor: | Predicted protein | | Authors: | Suskiewicz, M.J, Ahel, I. | | Deposit date: | 2020-01-13 | | Release date: | 2020-02-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.091 Å) | | Cite: | HPF1 completes the PARP active site for DNA damage-induced ADP-ribosylation.
Nature, 579, 2020
|
|
6TX2
 
 | | Human HPF1 | | Descriptor: | Histone PARylation factor 1, SODIUM ION | | Authors: | Suskiewicz, M.J, Ahel, I. | | Deposit date: | 2020-01-13 | | Release date: | 2020-02-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | HPF1 completes the PARP active site for DNA damage-induced ADP-ribosylation.
Nature, 579, 2020
|
|
6TX3
 
 | | HPF1 bound to catalytic fragment of PARP2 | | Descriptor: | 2-[4-[(2S,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]carbonylpiperazin-1-yl]-N-(1-oxidanylidene-2,3-dihydroisoindol-4-yl)ethanamide, Histone PARylation factor 1, Poly [ADP-ribose] polymerase 2,Poly [ADP-ribose] polymerase 2 | | Authors: | Suskiewicz, M.J, Ahel, I. | | Deposit date: | 2020-01-13 | | Release date: | 2020-02-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | HPF1 completes the PARP active site for DNA damage-induced ADP-ribosylation.
Nature, 579, 2020
|
|
6TV6
 
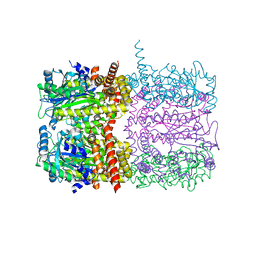 | | Octameric McsB from Bacillus subtilis. | | Descriptor: | MAGNESIUM ION, Protein-arginine kinase | | Authors: | Suskiewicz, M.J, Hajdusits, B, Meinhart, A, Clausen, T. | | Deposit date: | 2020-01-09 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | McsB forms a gated kinase chamber to mark aberrant bacterial proteins for degradation.
Elife, 10, 2021
|
|
8ODR
 
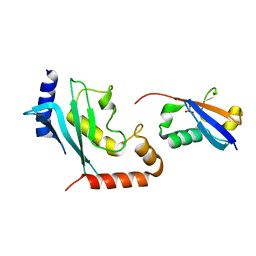 | | Mimetic of UBC9-SUMO1 | | Descriptor: | SULFATE ION, SUMO-conjugating enzyme UBC9, Small ubiquitin-related modifier 1 | | Authors: | Coste, F, Goffinont, S, Suskiewicz, M.J. | | Deposit date: | 2023-03-09 | | Release date: | 2023-06-07 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural insights into the regulation of the human E2∼SUMO conjugate through analysis of its stable mimetic.
J.Biol.Chem., 299, 2023
|
|
8P2O
 
 | |
8P2P
 
 | |
8P2N
 
 | |
8RIR
 
 | |
8RIT
 
 | |
