7CI9
 
 | | Crystal structure of P.aeruginosa LpxC in complex with inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-azanyl-2-[[4-[2-[3-[[2-[(1S)-1-oxidanylethyl]imidazol-1-yl]methyl]-1,2-oxazol-5-yl]ethynyl]phenoxy]methyl]propane-1,3-diol, PHOSPHATE ION, ... | | Authors: | Mima, M, Baker, L.M, Surgenor, A, Robertson, A. | | Deposit date: | 2020-07-07 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Fragment-Based Discovery of Novel Non-Hydroxamate LpxC Inhibitors with Antibacterial Activity.
J.Med.Chem., 63, 2020
|
|
7CI5
 
 | | Crystal structure of P.aeruginosa LpxC in complex with inhibitor | | Descriptor: | (3R)-3-azanyl-4-oxidanylidene-4-[[3-(trifluoromethyloxy)phenyl]amino]butanoic acid, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ... | | Authors: | Mima, M, Baker, L.M, Surgenor, A, Robertson, A. | | Deposit date: | 2020-07-07 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Fragment-Based Discovery of Novel Non-Hydroxamate LpxC Inhibitors with Antibacterial Activity.
J.Med.Chem., 63, 2020
|
|
7CIB
 
 | | Crystal structure of P.aeruginosa LpxC in complex with inhibitor | | Descriptor: | 2-oxidanyl-4-phenyl-benzoic acid, DIMETHYL SULFOXIDE, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ... | | Authors: | Baker, L.M, Mima, M, Surgenor, A, Robertson, A. | | Deposit date: | 2020-07-07 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Fragment-Based Discovery of Novel Non-Hydroxamate LpxC Inhibitors with Antibacterial Activity.
J.Med.Chem., 63, 2020
|
|
7CI7
 
 | | Crystal structure of P.aeruginosa LpxC in complex with inhibitor | | Descriptor: | (2R,3R)-2-azanyl-1-[4-[[4-[2-[4-(hydroxymethyl)phenyl]ethynyl]phenyl]methyl]piperidin-1-yl]-4-methylsulfonyl-3-oxidanyl-butan-1-one, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ZINC ION | | Authors: | Mima, M, Baker, L.M, Surgenor, A, Robertson, A. | | Deposit date: | 2020-07-07 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Fragment-Based Discovery of Novel Non-Hydroxamate LpxC Inhibitors with Antibacterial Activity.
J.Med.Chem., 63, 2020
|
|
7CI8
 
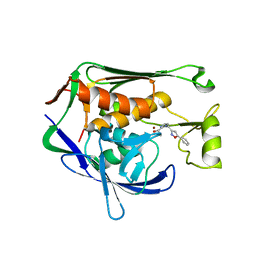 | | Crystal structure of P.aeruginosa LpxC in complex with inhibitor | | Descriptor: | (1S)-1-[1-[(5-phenyl-1,2-oxazol-3-yl)methyl]imidazol-2-yl]ethanol, MAGNESIUM ION, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ... | | Authors: | Mima, M, Baker, L.M, Surgenor, A, Robertson, A. | | Deposit date: | 2020-07-07 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Fragment-Based Discovery of Novel Non-Hydroxamate LpxC Inhibitors with Antibacterial Activity.
J.Med.Chem., 63, 2020
|
|
7CIE
 
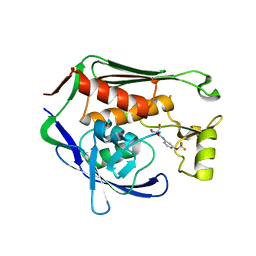 | | Crystal structure of P.aeruginosa LpxC in complex with inhibitor | | Descriptor: | (2R)-2-azanyl-3-oxidanyl-N-[3-(trifluoromethyloxy)phenyl]propanamide, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ZINC ION | | Authors: | Baker, L.M, Mima, M, Surgenor, A, Robertson, A. | | Deposit date: | 2020-07-07 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Fragment-Based Discovery of Novel Non-Hydroxamate LpxC Inhibitors with Antibacterial Activity.
J.Med.Chem., 63, 2020
|
|
7CI6
 
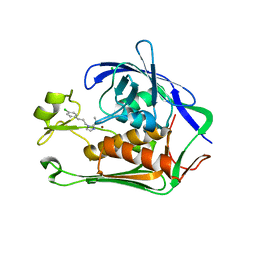 | | Crystal structure of P.aeruginosa LpxC in complex with inhibitor | | Descriptor: | (1S)-1-[1-[3-(4-chlorophenyl)propyl]imidazol-2-yl]ethanol, CHLORIDE ION, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ... | | Authors: | Mima, M, Baker, L.M, Surgenor, A, Robertson, A. | | Deposit date: | 2020-07-07 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Fragment-Based Discovery of Novel Non-Hydroxamate LpxC Inhibitors with Antibacterial Activity.
J.Med.Chem., 63, 2020
|
|
7CIA
 
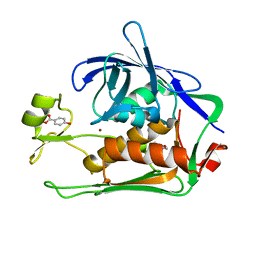 | | Crystal structure of P.aeruginosa LpxC in complex with inhibitor | | Descriptor: | 4-HYDROXY-BENZOIC ACID METHYL ESTER, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ZINC ION | | Authors: | Baker, L.M, Mima, M, Surgenor, A, Robertson, A. | | Deposit date: | 2020-07-07 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Fragment-Based Discovery of Novel Non-Hydroxamate LpxC Inhibitors with Antibacterial Activity.
J.Med.Chem., 63, 2020
|
|
2BRB
 
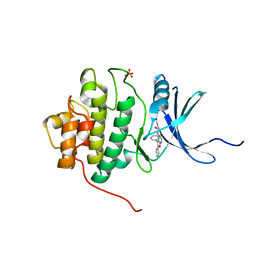 | | Structure-based Design of Novel Chk1 Inhibitors: Insights into Hydrogen Bonding and Protein-Ligand Affinity | | Descriptor: | 2-[(5,6-DIPHENYLFURO[2,3-D]PYRIMIDIN-4-YL)AMINO]ETHANOL, SERINE/THREONINE-PROTEIN KINASE CHK1, SULFATE ION | | Authors: | Foloppe, N, Fisher, L.M, Howes, R, Kierstan, P, Potter, A, Robertson, A.G.S, Surgenor, A.E. | | Deposit date: | 2005-05-04 | | Release date: | 2005-05-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Based Design of Novel Chk1 Inhibitors: Insights Into Hydrogen Bonding and Protein-Ligand Affinity.
J.Med.Chem., 48, 2005
|
|
2BRH
 
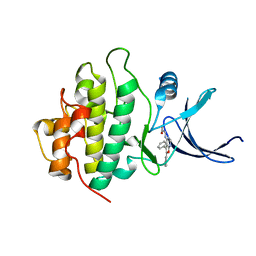 | | Structure-based Design of Novel Chk1 Inhibitors: Insights into Hydrogen Bonding and Protein-Ligand Affinity | | Descriptor: | N-(5,6-DIPHENYLFURO[2,3-D]PYRIMIDIN-4-YL)GLYCINE, SERINE/THREONINE-PROTEIN KINASE CHK1 | | Authors: | Foloppe, N, Fisher, L.M, Howes, R, Kierstan, P, Potter, A, Robertson, A.G.S, Surgenor, A.E. | | Deposit date: | 2005-05-05 | | Release date: | 2005-05-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Based Design of Novel Chk1 Inhibitors: Insights Into Hydrogen Bonding and Protein-Ligand Affinity.
J.Med.Chem., 48, 2005
|
|
2BRG
 
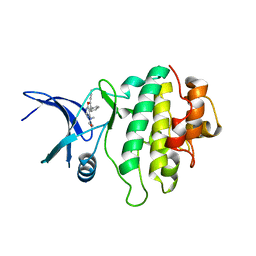 | | Structure-based Design of Novel Chk1 Inhibitors: Insights into Hydrogen Bonding and Protein-Ligand Affinity | | Descriptor: | (5,6-DIPHENYL-FURO[2,3-D]PYRIMIDIN-4-YLAMINO)-ACETIC, SERINE/THREONINE-PROTEIN KINASE CHK1 | | Authors: | Foloppe, N, Fisher, L.M, Howes, R, Kierstan, P, Potter, A, Robertson, A.G.S, Surgenor, A.E. | | Deposit date: | 2005-05-05 | | Release date: | 2005-05-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Based Design of Novel Chk1 Inhibitors: Insights Into Hydrogen Bonding and Protein-Ligand Affinity.
J.Med.Chem., 48, 2005
|
|
2BRM
 
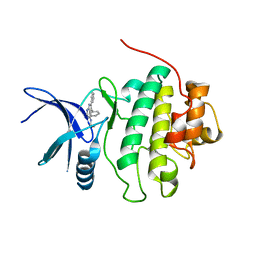 | | Structure-based Design of Novel Chk1 Inhibitors: Insights into Hydrogen Bonding and Protein-Ligand Affinity | | Descriptor: | 3-AMINO-3-BENZYL-[4.3.0]BICYCLO-1,6-DIAZANONAN-2-ONE, SERINE/THREONINE-PROTEIN KINASE CHK1 | | Authors: | Foloppe, N, Fisher, L.M, Howes, R, Kierstan, P, Potter, A, Robertson, A.G.S, Surgenor, A.E. | | Deposit date: | 2005-05-09 | | Release date: | 2005-05-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Based Design of Novel Chk1 Inhibitors: Insights Into Hydrogen Bonding and Protein-Ligand Affinity.
J.Med.Chem., 48, 2005
|
|
2BRN
 
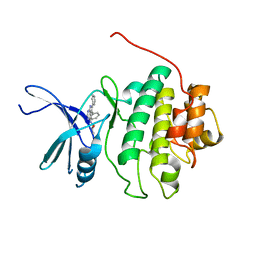 | | Structure-based Design of Novel Chk1 Inhibitors: Insights into Hydrogen Bonding and Protein-Ligand Affinity | | Descriptor: | (2R)-1-[(5,6-DIPHENYL-7H-PYRROLO[2,3-D]PYRIMIDIN-4-YL)AMINO]PROPAN-2-OL, SERINE/THREONINE-PROTEIN KINASE CHK1 | | Authors: | Foloppe, N, Fisher, L.M, Howes, R, Kierstan, P, Potter, A, Robertson, A.G.S, Surgenor, A.E. | | Deposit date: | 2005-05-09 | | Release date: | 2005-05-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-Based Design of Novel Chk1 Inhibitors: Insights Into Hydrogen Bonding and Protein-Ligand Affinity.
J.Med.Chem., 48, 2005
|
|
2BR1
 
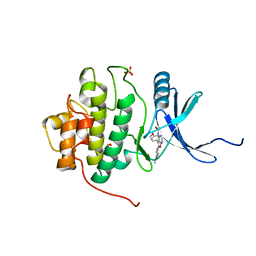 | | Structure-based Design of Novel Chk1 Inhibitors: Insights into Hydrogen Bonding and Protein-Ligand Affinity | | Descriptor: | 2-[5,6-BIS-(4-METHOXY-PHENYL)-FURO[2,3-D]PYRIMIDIN-4-YLAMINO]-ETHANOL, SERINE/THREONINE-PROTEIN KINASE CHK1, SULFATE ION | | Authors: | Foloppe, N, Fisher, L.M, Howes, R, Kierstan, P, Potter, A, Robertson, A.G.S, Surgenor, A.E. | | Deposit date: | 2005-04-29 | | Release date: | 2005-05-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Based Design of Novel Chk1 Inhibitors: Insights Into Hydrogen Bonding and Protein-Ligand Affinity.
J.Med.Chem., 48, 2005
|
|
2BRO
 
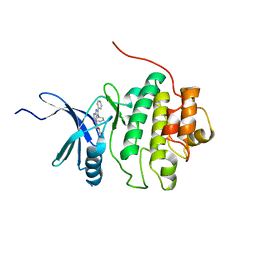 | | Structure-based Design of Novel Chk1 Inhibitors: Insights into Hydrogen Bonding and Protein-Ligand Affinity | | Descriptor: | (2R)-3-{[(4Z)-5,6-DIPHENYL-6,7-DIHYDRO-4H-PYRROLO[2,3-D]PYRIMIDIN-4-YLIDENE]AMINO}PROPANE-1,2-DIOL, SERINE/THREONINE-PROTEIN KINASE CHK1 | | Authors: | Foloppe, N, Fisher, L.M, Howes, R, Kierstan, P, Potter, A, Robertson, A.G.S, Surgenor, A.E. | | Deposit date: | 2005-05-09 | | Release date: | 2005-05-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Based Design of Novel Chk1 Inhibitors: Insights Into Hydrogen Bonding and Protein-Ligand Affinity.
J.Med.Chem., 48, 2005
|
|
2CCS
 
 | | HUMAN HSP90 WITH 4-CHLORO-6-(4-PIPERAZIN-1-YL-1H-PYRAZOL-3-YL)- BENZENE-1,2-DIOL | | Descriptor: | 4-CHLORO-6-(4-PIPERAZIN-1-YL-1H-PYRAZOL-5-YL)BENZENE-1,3-DIOL, HEAT SHOCK PROTEIN HSP-90 ALPHA | | Authors: | Barril, X, Beswick, M.C, Collier, A, Drysdale, M.J, Dymock, B.W, Fink, A, Grant, K, Howes, R, Jordan, A.M, Massey, A, Surgenor, A, Wayne, J, Workman, P, Wright, L. | | Deposit date: | 2006-01-18 | | Release date: | 2006-02-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | 4-Amino derivatives of the Hsp90 inhibitor CCT018159.
Bioorg. Med. Chem. Lett., 16, 2006
|
|
2CCT
 
 | | HUMAN HSP90 WITH 5-(5-CHLORO-2,4-DIHYDROXY-PHENYL)-4-PIPERAZIN-1-YL- 2H-PYRAZOLE-3-CARBOXYLIC ACID ETHYLAMIDE | | Descriptor: | 5-(5-CHLORO-2,4-DIHYDROXYPHENYL)-N-ETHYL-4-PIPERAZIN-1-YL-1H-PYRAZOLE-3-CARBOXAMIDE, HEAT SHOCK PROTEIN HSP-90 ALPHA | | Authors: | Barril, X, Beswick, M.C, Collier, A, Drysdale, M.J, Dymock, B.W, Fink, A, Grant, K, Howes, R, Jordan, A.M, Massey, A, Surgenor, A, Wayne, J, Workman, P, Wright, L. | | Deposit date: | 2006-01-18 | | Release date: | 2006-02-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | 4-Amino derivatives of the Hsp90 inhibitor CCT018159.
Bioorg. Med. Chem. Lett., 16, 2006
|
|
2CCU
 
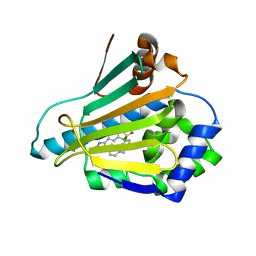 | | HUMAN HSP90 WITH 4-CHLORO-6-(4-(4-(4-METHANESULPHONYL-BENZYL)- PIERAZIN-1-YL)-1H-PYRAZOL-3-YL)-BENZENE-1,3-DIOL | | Descriptor: | 4-CHLORO-6-(4-{4-[4-(METHYLSULFONYL)BENZYL]PIPERAZIN-1-YL}-1H-PYRAZOL-5-YL)BENZENE-1,3-DIOL, HEAT SHOCK PROTEIN HSP-90 ALPHA | | Authors: | Barril, X, Beswick, M.C, Collier, A, Drysdale, M.J, Dymock, B.W, Fink, A, Grant, K, Howes, R, Jordan, A.M, Massey, A, Surgenor, A, Wayne, J, Workman, P, Wright, L. | | Deposit date: | 2006-01-18 | | Release date: | 2006-02-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | 4-Amino derivatives of the Hsp90 inhibitor CCT018159.
Bioorg. Med. Chem. Lett., 16, 2006
|
|
2BT0
 
 | | Novel, potent small molecule inhibitors of the molecular chaperone Hsp90 discovered through structure-based design | | Descriptor: | 4-[4-(2,3-DIHYDRO-1,4-BENZODIOXIN-6-YL)-3-METHYL-1H-PYRAZOL-5-YL]-6-ETHYLBENZENE-1,3-DIOL, HEAT SHOCK PROTEIN HSP90-ALPHA | | Authors: | Dymock, B.W, Barril, X, Brough, P.A, Cansfield, J.E, Massey, A, McDonald, E, Hubbard, R.E, Surgenor, A, Roughley, S.D, Webb, P, Workman, P, Wright, L, Drysdale, M.J. | | Deposit date: | 2005-05-24 | | Release date: | 2005-06-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Novel, potent small-molecule inhibitors of the molecular chaperone Hsp90 discovered through structure-based design.
J. Med. Chem., 48, 2005
|
|
2BZ5
 
 | | Structure-based Discovery of a New Class of Hsp90 Inhibitors | | Descriptor: | 2,5-DICHLORO-N-[4-HYDROXY-3-(2-HYDROXY-1-NAPHTHYL)PHENYL]BENZENESULFONAMIDE, HEAT SHOCK PROTEIN HSP90-ALPHA | | Authors: | Barril, X, Brough, P, Drysdale, M, Hubbard, R.E, Massey, A, Surgenor, A, Wright, L. | | Deposit date: | 2005-08-11 | | Release date: | 2005-10-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Based Discovery of a New Class of Hsp90 Inhibitors.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
7A4O
 
 | | Structure of DYRK1A in complex with AMPNP | | Descriptor: | Dual specificity tyrosine-phosphorylation-regulated kinase 1A, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Dokurno, P, Surgenor, A.E, Hubbard, R.E. | | Deposit date: | 2020-08-20 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Fragment-Derived Selective Inhibitors of Dual-Specificity Kinases DYRK1A and DYRK1B.
J.Med.Chem., 64, 2021
|
|
7A53
 
 | | Structure of DYRK1A in complex with compound 7 | | Descriptor: | (3~{E})-3-[(5-methylfuran-2-yl)methylidene]-1~{H}-indol-2-one, Dual specificity tyrosine-phosphorylation-regulated kinase 1A | | Authors: | Dokurno, P, Surgenor, A.E, Hubbard, R.E. | | Deposit date: | 2020-08-20 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Fragment-Derived Selective Inhibitors of Dual-Specificity Kinases DYRK1A and DYRK1B.
J.Med.Chem., 64, 2021
|
|
7A4R
 
 | | Structure of DYRK1A in complex with compound 1 | | Descriptor: | 2-amino-3H,4H,7H-pyrrolo[2,3-d]pyrimidin-4-one, DIMETHYL SULFOXIDE, Dual specificity tyrosine-phosphorylation-regulated kinase 1A | | Authors: | Dokurno, P, Surgenor, A.E, Hubbard, R.E. | | Deposit date: | 2020-08-20 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Fragment-Derived Selective Inhibitors of Dual-Specificity Kinases DYRK1A and DYRK1B.
J.Med.Chem., 64, 2021
|
|
7A4S
 
 | | Structure of DYRK1A in complex with compound 2 | | Descriptor: | 7-chlorothieno[3,2-c]pyridin-4-amine, Dual specificity tyrosine-phosphorylation-regulated kinase 1A | | Authors: | Dokurno, P, Surgenor, A.E, Hubbard, R.E. | | Deposit date: | 2020-08-20 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Fragment-Derived Selective Inhibitors of Dual-Specificity Kinases DYRK1A and DYRK1B.
J.Med.Chem., 64, 2021
|
|
7A4Z
 
 | | Structure of DYRK1A in complex with compound 4 | | Descriptor: | 3-(4-methoxyphenyl)-1~{H}-pyrazol-5-amine, Dual specificity tyrosine-phosphorylation-regulated kinase 1A | | Authors: | Dokurno, P, Surgenor, A.E, Hubbard, R.E. | | Deposit date: | 2020-08-20 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Fragment-Derived Selective Inhibitors of Dual-Specificity Kinases DYRK1A and DYRK1B.
J.Med.Chem., 64, 2021
|
|
