1I33
 
 | | LEISHMANIA MEXICANA GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE IN COMPLEX WITH INHIBITORS | | Descriptor: | GLYCERALDEHYDE 3-PHOSPHATE DEHYDROGENASE, N-1,2,3,4-TETRAHYDRONAPHTH-1-YL-2'-[3,5-DIMETHOXYBENZAMIDO]-2'-DEOXY-ADENOSINE | | Authors: | Suresh, S, Bressi, J.C, Kennedy, K.J, Verlinde, C.L.M.J, Gelb, M.H, Hol, W.G.J. | | Deposit date: | 2001-02-12 | | Release date: | 2001-10-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Conformational changes in Leishmania mexicana glyceraldehyde-3-phosphate dehydrogenase induced by designed inhibitors.
J.Mol.Biol., 309, 2001
|
|
1I32
 
 | | LEISHMANIA MEXICANA GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE IN COMPLEX WITH INHIBITORS | | Descriptor: | GLYCERALDEHYDE 3-PHOSPHATE DEHYDROGENASE, N-NAPHTHALEN-1-YLMETHYL-2'-[3,5-DIMETHOXYBENZAMIDO]-2'-DEOXY-ADENOSINE | | Authors: | Suresh, S, Bressi, J.C, Kennedy, K.J, Verlinde, C.L.M.J, Gelb, M.H, Hol, W.G.J. | | Deposit date: | 2001-02-12 | | Release date: | 2001-10-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Conformational changes in Leishmania mexicana glyceraldehyde-3-phosphate dehydrogenase induced by designed inhibitors.
J.Mol.Biol., 309, 2001
|
|
1JDJ
 
 | | CRYSTAL STRUCTURE OF LEISHMANIA MEXICANA GLYCEROL-3-PHOSPHATE DEHYDROGENASE IN COMPLEX WITH 2-FLUORO-6-CHLOROPURINE | | Descriptor: | 6-CHLORO-2-FLUOROPURINE, GLYCEROL-3-PHOSPHATE DEHYDROGENASE, PENTADECANE | | Authors: | Suresh, S, Wisedchaisri, G, Kennedy, K.J, Verlinde, C.L.M.J, Gelb, M.H, Hol, W.G.J. | | Deposit date: | 2001-06-14 | | Release date: | 2002-06-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Anomalous differences of light elements in determining precise binding modes of ligands to glycerol-3-phosphate dehydrogenase.
Chem.Biol., 9, 2002
|
|
1EVZ
 
 | | CRYSTAL STRUCTURE OF LEISHMANIA MEXICANA GLYCEROL-3-PHOSPHATE DEHYDROGENASE IN COMPLEX WITH NAD | | Descriptor: | GLYCEROL-3-PHOSPHATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PENTADECANE | | Authors: | Suresh, S, Turley, S, Opperdoes, F.R, Michels, P.A.M, Hol, W.G.J. | | Deposit date: | 2000-04-21 | | Release date: | 2001-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A potential target enzyme for trypanocidal drugs revealed by the crystal structure of NAD-dependent glycerol-3-phosphate dehydrogenase from Leishmania mexicana.
Structure Fold.Des., 8, 2000
|
|
1EVY
 
 | | CRYSTAL STRUCTURE OF LEISHMANIA MEXICANA GLYCEROL-3-PHOSPHATE DEHYDROGENASE | | Descriptor: | GLYCEROL-3-PHOSPHATE DEHYDROGENASE, PENTADECANE | | Authors: | Suresh, S, Turley, S, Opperdoes, F.R, Michels, P.A.M, Hol, W.G.J. | | Deposit date: | 2000-04-20 | | Release date: | 2001-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A potential target enzyme for trypanocidal drugs revealed by the crystal structure of NAD-dependent glycerol-3-phosphate dehydrogenase from Leishmania mexicana.
Structure Fold.Des., 8, 2000
|
|
1GYQ
 
 | |
9B9G
 
 | | Structure of the PI4KA complex bound to Calcineurin | | Descriptor: | CALCIUM ION, Calcineurin subunit B type 1, Hyccin, ... | | Authors: | Shaw, A.L, Suresh, S, Yip, C.K, Burke, J.E. | | Deposit date: | 2024-04-02 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of calcineurin bound to PI4KA reveals dual interface in both PI4KA and FAM126A.
Structure, 2024
|
|
1M66
 
 | | Crystal Structure of Leishmania mexicana GPDH Complexed with Inhibitor 2-bromo-6-chloro-purine | | Descriptor: | 2-BROMO-6-CHLORO-PURINE, Glycerol-3-phosphate dehydrogenase, PALMITIC ACID | | Authors: | Choe, J, Suresh, S, Wisedchaisri, G, Kennedy, K.J, Gelb, M.H, Hol, W.G.J. | | Deposit date: | 2002-07-12 | | Release date: | 2002-12-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Anomalous differences of light elements in determining precise binding modes of ligands to
glycerol-3-phosphate dehydrogenase
Chem.Biol., 9, 2002
|
|
1M67
 
 | | Crystal Structure of Leishmania mexicana GPDH Complexed with Inhibitor 2-bromo-6-hydroxy-purine | | Descriptor: | 2-BROMO-6-HYDROXY-PURINE, Glycerol-3-phosphate dehydrogenase, PALMITIC ACID | | Authors: | Choe, J, Suresh, S, Wisedchaisri, G, Kennedy, K.J, Gelb, M.H, Hol, W.G.J. | | Deposit date: | 2002-07-12 | | Release date: | 2002-12-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Anomalous differences of light elements in determining precise binding modes of ligands to
glycerol-3-phosphate dehydrogenase
Chem.Biol., 9, 2002
|
|
8D3Q
 
 | | Type I-C Cas4-Cas1-Cas2 complex bound to a PAM/NoPAM prespacer | | Descriptor: | CRISPR-associated endonuclease Cas1, CRISPR-associated endonuclease Cas2, CRISPR-associated exonuclease Cas4, ... | | Authors: | Dhingra, Y, Suresh, S.K, Juneja, P, Sashital, D.G. | | Deposit date: | 2022-06-01 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | PAM binding ensures orientational integration during Cas4-Cas1-Cas2-mediated CRISPR adaptation.
Mol.Cell, 82, 2022
|
|
8D3M
 
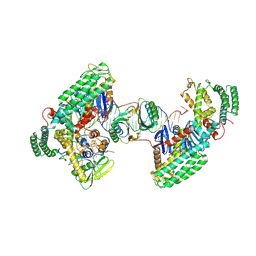 | | Type I-C Cas4-Cas1-Cas2 complex bound to a PAM/Processed prespacer | | Descriptor: | CRISPR-associated endonuclease Cas1, CRISPR-associated endonuclease Cas2, CRISPR-associated exonuclease Cas4, ... | | Authors: | Dhingra, Y, Suresh, S.K, Juneja, P, Sashital, D.G. | | Deposit date: | 2022-06-01 | | Release date: | 2022-11-02 | | Last modified: | 2022-11-30 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | PAM binding ensures orientational integration during Cas4-Cas1-Cas2-mediated CRISPR adaptation.
Mol.Cell, 82, 2022
|
|
8D3L
 
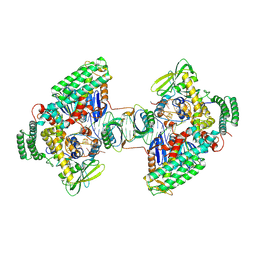 | | Type I-C Cas4-Cas1-Cas2 complex bound to a PAM/PAM prespacer | | Descriptor: | CRISPR-associated endonuclease Cas1, CRISPR-associated endonuclease Cas2, CRISPR-associated exonuclease Cas4, ... | | Authors: | Dhingra, Y, Suresh, S.K, Juneja, P, Sashital, D.G. | | Deposit date: | 2022-06-01 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | PAM binding ensures orientational integration during Cas4-Cas1-Cas2-mediated CRISPR adaptation.
Mol.Cell, 82, 2022
|
|
8D3P
 
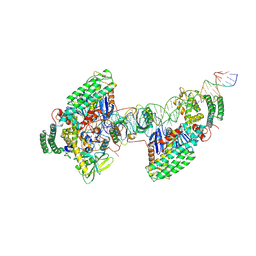 | | Type I-C Cas4-Cas1-Cas2 complex bound to half-site integration intermediate (HSI) | | Descriptor: | CRISPR-associated endonuclease Cas1, CRISPR-associated endonuclease Cas2, CRISPR-associated exonuclease Cas4, ... | | Authors: | Dhingra, Y, Suresh, S.K, Juneja, P, Sashital, D.G. | | Deposit date: | 2022-06-01 | | Release date: | 2022-11-02 | | Last modified: | 2022-11-30 | | Method: | ELECTRON MICROSCOPY (4.26 Å) | | Cite: | PAM binding ensures orientational integration during Cas4-Cas1-Cas2-mediated CRISPR adaptation.
Mol.Cell, 82, 2022
|
|
