1H65
 
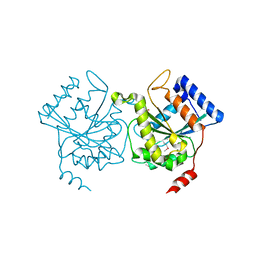 | | Crystal structure of pea Toc34 - a novel GTPase of the chloroplast protein translocon | | Descriptor: | CHLOROPLAST OUTER ENVELOPE PROTEIN OEP34, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Sun, Y.J, Forouhar, F, Li, H.M, Tu, S.L, Kao, S, Shr, H.L, Chou, C.C, Hsiao, C.D. | | Deposit date: | 2001-06-06 | | Release date: | 2002-01-29 | | Last modified: | 2019-06-12 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Pea Toc34 - a Novel Gtpase of the Chloroplast Protein Translocon
Nat.Struct.Biol., 9, 2002
|
|
5EZ1
 
 | |
6MYE
 
 | | Crystal structure of human Scribble PDZ1 domain in complex with internal PDZ binding motif of Src homology 3 domain-containing guanine nucleotide exchange factor (SGEF) | | Descriptor: | FORMIC ACID, Protein scribble homolog, Rho guanine nucleotide exchange factor 26, ... | | Authors: | Sun, Y.J, Hou, T, Gakhar, L, Fuentes, E.J. | | Deposit date: | 2018-11-01 | | Release date: | 2019-04-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | SGEF forms a complex with Scribble and Dlg1 and regulates epithelial junctions and contractility.
J.Cell Biol., 218, 2019
|
|
6MYF
 
 | |
6NID
 
 | |
6NEK
 
 | |
6NH9
 
 | |
7CJQ
 
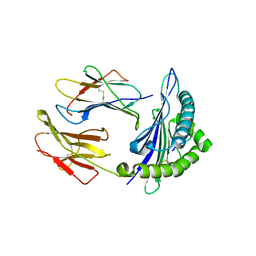 | | Structure of DLA-88*001:04 | | Descriptor: | ARG-THR-ILE-SER-TYR-THR-TYR-PRO-PHE, Beta-2-microglobulin, MHC class I DLA-88 | | Authors: | Sun, Y.J, Ma, L.Z, Li, S. | | Deposit date: | 2020-07-12 | | Release date: | 2020-08-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of canine MHC class I at 2.7 Angstroms resolution
To Be Published
|
|
2PV6
 
 | | HIV-1 gp41 Membrane Proximal Ectodomain Region peptide in DPC micelle | | Descriptor: | Envelope glycoprotein | | Authors: | Sun, Z.-Y.J, Oh, K.J, Kim, M, Reinherz, E.L, Wagner, G. | | Deposit date: | 2007-05-09 | | Release date: | 2008-03-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | HIV-1 broadly neutralizing antibody extracts its epitope from a kinked gp41 ectodomain region on the viral membrane
Immunity, 28, 2008
|
|
1CI5
 
 | | GLYCAN-FREE MUTANT ADHESION DOMAIN OF HUMAN CD58 (LFA-3) | | Descriptor: | PROTEIN (LYMPHOCYTE FUNCTION-ASSOCIATED ANTIGEN 3(CD58)) | | Authors: | Sun, Z.Y.J, Dotsch, V, Kim, M, Li, J, Reinherz, E.L, Wagner, G. | | Deposit date: | 1999-04-07 | | Release date: | 1999-06-22 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Functional glycan-free adhesion domain of human cell surface receptor CD58: design, production and NMR studies.
EMBO J., 18, 1999
|
|
1JBJ
 
 | | CD3 Epsilon and gamma Ectodomain Fragment Complex in Single-Chain Construct | | Descriptor: | CD3 Epsilon and gamma Ectodomain Fragment Complex | | Authors: | Sun, Z.-Y.J, Kim, K.S, Wagner, G, Reinherz, E.L. | | Deposit date: | 2001-06-05 | | Release date: | 2001-12-05 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Mechanisms contributing to T cell receptor signaling and assembly revealed by the solution structure of an ectodomain fragment of the CD3 epsilon gamma heterodimer.
Cell(Cambridge,Mass.), 105, 2001
|
|
1XMW
 
 | | CD3 EPSILON AND DELTA ECTODOMAIN FRAGMENT COMPLEX IN SINGLE-CHAIN CONSTRUCT | | Descriptor: | Chimeric CD3 mouse Epsilon and sheep Delta Ectodomain Fragment Complex | | Authors: | Sun, Z.-Y.J, Kim, S.T, Kim, I.C, Fahmy, A, Reinherz, E.L, Wagner, G. | | Deposit date: | 2004-10-04 | | Release date: | 2004-11-30 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the CD3epsilondelta ectodomain and comparison with CD3epsilongamma as a basis for modeling T cell receptor topology and signaling.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
5IJ4
 
 | | Solution structure of AN1-type zinc finger domain from Cuz1 (Cdc48 associated ubiquitin-like/zinc-finger protein-1) | | Descriptor: | CDC48-associated ubiquitin-like/zinc finger protein 1, ZINC ION | | Authors: | Sun, Z.-Y.J, Hanna, J, Wagner, G, Bhanu, M.K, Allan, M, Arthanari, H. | | Deposit date: | 2016-03-01 | | Release date: | 2016-10-05 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Cuz1 AN1 Zinc Finger Domain: An Exposed LDFLP Motif Defines a Subfamily of AN1 Proteins.
Plos One, 11, 2016
|
|
4UMK
 
 | | The complex of Spo0J and parS DNA in chromosomal partition system | | Descriptor: | DNA, PROBABLE CHROMOSOME-PARTITIONING PROTEIN PARB, SULFATE ION | | Authors: | Chen, B.W, Chu, C.H, Tung, J.Y, Hsu, C.E, Hsiao, C.D, Sun, Y.J. | | Deposit date: | 2014-05-19 | | Release date: | 2015-05-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.096 Å) | | Cite: | Insights into ParB spreading from the complex structure of Spo0J and parS.
Proc. Natl. Acad. Sci. U.S.A., 112, 2015
|
|
1E9L
 
 | |
6P3Q
 
 | | Calpain-5 (CAPN5) Protease Core (PC) | | Descriptor: | Calpain-5 | | Authors: | Velez, G, Sun, Y.J, Khan, S, Yang, J, Koster, H.J, Lokesh, G, Mahajan, V. | | Deposit date: | 2019-05-24 | | Release date: | 2020-02-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Insights into the Unique Activation Mechanisms of a Non-classical Calpain and Its Disease-Causing Variants.
Cell Rep, 30, 2020
|
|
4A7W
 
 | | Crystal structure of uridylate kinase from Helicobacter pylori | | Descriptor: | GLYCEROL, GUANOSINE-5'-TRIPHOSPHATE, URIDYLATE KINASE | | Authors: | Chu, C.H, Chen, P.C, Liu, M.H, Sun, Y.J. | | Deposit date: | 2011-11-15 | | Release date: | 2012-06-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of Helicobacter Pylori Uridylate Kinase: Insight Into Release of the Product Udp
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4A7X
 
 | | Crystal structure of uridylate kinase from Helicobacter pylori | | Descriptor: | URIDINE-5'-DIPHOSPHATE, URIDYLATE KINASE | | Authors: | Chu, C.H, Liu, M.H, Chen, P.C, Sun, Y.J. | | Deposit date: | 2011-11-15 | | Release date: | 2012-06-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structures of Helicobacter Pylori Uridylate Kinase: Insight Into Release of the Product Udp
Acta Crystallogr.,Sect.D, 68, 2012
|
|
2W58
 
 | | Crystal Structure of the DnaI | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Tsai, K.L, Lo, Y.H, Sun, Y.J, Hsiao, C.D. | | Deposit date: | 2008-12-08 | | Release date: | 2009-12-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular Interplay between the Replicative Helicase Dnac and its Loader Protein Dnai from Geobacillus Kaustophilus.
J.Mol.Biol., 393, 2009
|
|
2VYE
 
 | | Crystal Structure of the DnaC-ssDNA complex | | Descriptor: | 5'-D(*TP*TP*TP*TP*TP*TP*TP*TP*TP)-3', REPLICATIVE DNA HELICASE | | Authors: | Lo, Y.H, Tsai, K.L, Sun, Y.J, Hsiao, C.D. | | Deposit date: | 2008-07-23 | | Release date: | 2008-12-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | The Crystal Structure of a Replicative Hexameric Helicase Dnac and its Complex with Single-Stranded DNA.
Nucleic Acids Res., 37, 2009
|
|
2VYF
 
 | | Crystal Structure of the DnaC | | Descriptor: | GOLD ION, REPLICATIVE DNA HELICASE | | Authors: | Lo, Y.H, Tsai, K.L, Sun, Y.J, Hsiao, C.D. | | Deposit date: | 2008-07-23 | | Release date: | 2008-12-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | The Crystal Structure of a Replicative Hexameric Helicase Dnac and its Complex with Single-Stranded DNA.
Nucleic Acids Res., 37, 2009
|
|
8YK9
 
 | | Structure of Saccharolobus solfataricus SegC (SSO0033) protein, ATP soak | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, SegC | | Authors: | Yen, C.Y, Lin, M.G, Sun, Y.J, Hsiao, C.D. | | Deposit date: | 2024-03-04 | | Release date: | 2024-08-07 | | Last modified: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Unraveling the structure and function of a novel SegC protein interacting with the SegAB chromosome segregation complex in Archaea.
Nucleic Acids Res., 52, 2024
|
|
6LX0
 
 | |
4BFN
 
 | | Crystal Structure of the Starch-Binding Domain from Rhizopus oryzae Glucoamylase in Complex with isomaltotetraose | | Descriptor: | GLUCOAMYLASE, alpha-D-glucopyranose-(1-6)-alpha-D-glucopyranose-(1-6)-alpha-D-glucopyranose-(1-6)-alpha-D-glucopyranose | | Authors: | Chu, C.H, Li, K.M, Lin, S.W, Sun, Y.J. | | Deposit date: | 2013-03-21 | | Release date: | 2013-10-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Crystal Structures of Starch Binding Domain from Rhizopus Oryzae Glucoamylase in Complex with Isomaltooligosaccharide: Insights Into Polysaccharide Binding Mechanism of Cbm21 Family.
Proteins, 82, 2014
|
|
4BFO
 
 | | Crystal Structure of the Starch-Binding Domain from Rhizopus oryzae Glucoamylase in Complex with isomaltotriose | | Descriptor: | GLUCOAMYLASE, alpha-D-glucopyranose-(1-6)-alpha-D-glucopyranose-(1-6)-alpha-D-glucopyranose | | Authors: | Chu, C.H, Li, K.M, Lin, S.W, Sun, Y.J. | | Deposit date: | 2013-03-21 | | Release date: | 2013-10-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.175 Å) | | Cite: | Crystal Structures of Starch Binding Domain from Rhizopus Oryzae Glucoamylase in Complex with Isomaltooligosaccharide: Insights Into Polysaccharide Binding Mechanism of Cbm21 Family.
Proteins, 82, 2014
|
|
