1A13
 
 | | G PROTEIN-BOUND CONFORMATION OF MASTOPARAN-X, NMR, 14 STRUCTURES | | Descriptor: | MASTOPARAN-X | | Authors: | Kusunoki, H, Wakamatsu, K, Sato, K, Miyazawa, T, Kohno, T. | | Deposit date: | 1997-12-20 | | Release date: | 1999-01-13 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | G protein-bound conformation of mastoparan-X: heteronuclear multidimensional transferred nuclear overhauser effect analysis of peptide uniformly enriched with 13C and 15N.
Biochemistry, 37, 1998
|
|
1S35
 
 | |
1U4Q
 
 | | Crystal Structure of Repeats 15, 16 and 17 of Chicken Brain Alpha Spectrin | | Descriptor: | Spectrin alpha chain, brain | | Authors: | Kusunoki, H, Minasov, G, MacDonald, R.I, Mondragon, A. | | Deposit date: | 2004-07-26 | | Release date: | 2004-10-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Independent Movement, Dimerization and Stability of Tandem Repeats of Chicken Brain alpha-Spectrin
J.Mol.Biol., 344, 2004
|
|
1U5P
 
 | | Crystal Structure of Repeats 15 and 16 of Chicken Brain Alpha Spectrin | | Descriptor: | PHOSPHATE ION, POTASSIUM ION, Spectrin alpha chain, ... | | Authors: | Kusunoki, H, Minasov, G, MacDonald, R.I, Mondragon, A. | | Deposit date: | 2004-07-28 | | Release date: | 2004-10-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Independent Movement, Dimerization and Stability of Tandem Repeats of Chicken Brain alpha-Spectrin
J.Mol.Biol., 344, 2004
|
|
2UYG
 
 | |
7C7V
 
 | | Vitamin D3 receptor/lithochoric acid derivative complex | | Descriptor: | (4R)-4-[(3R,5R,8R,9S,10S,13R,14S,17R)-10,13-dimethyl-3-(2-methyl-2-oxidanyl-propyl)-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthren-17-yl]pentanoic acid, FORMIC ACID, Mediator of RNA polymerase II transcription subunit 1, ... | | Authors: | Masuno, H, Numoto, N, Kagechika, H, Ito, N. | | Deposit date: | 2020-05-26 | | Release date: | 2021-01-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Lithocholic Acid Derivatives as Potent Vitamin D Receptor Agonists.
J.Med.Chem., 64, 2021
|
|
7C7W
 
 | | Vitamin D3 receptor/lithochoric acid derivative complex | | Descriptor: | (4R)-4-[(3S,5R,8R,9S,10S,13R,14S,17R)-10,13-dimethyl-3-(2-methyl-2-oxidanyl-propyl)-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthren-17-yl]pentanoic acid, FORMIC ACID, Mediator of RNA polymerase II transcription subunit 1, ... | | Authors: | Masuno, H, Numoto, N, Kagechika, H, Ito, N. | | Deposit date: | 2020-05-26 | | Release date: | 2021-01-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Lithocholic Acid Derivatives as Potent Vitamin D Receptor Agonists.
J.Med.Chem., 64, 2021
|
|
6K5O
 
 | | Development of Novel Lithocholic Acid Derivatives as Vitamin D Receptor Agonists | | Descriptor: | (4~{R})-4-[(3~{R},5~{R},8~{R},9~{S},10~{S},13~{R},14~{S},17~{R})-10,13-dimethyl-3-methylsulfonyloxy-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1~{H}-cyclopenta[a]phenanthren-17-yl]pentanoic acid, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | Authors: | Masuno, H, Kagechika, H, Ito, N. | | Deposit date: | 2019-05-29 | | Release date: | 2019-07-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Development of novel lithocholic acid derivatives as vitamin D receptor agonists.
Bioorg.Med.Chem., 27, 2019
|
|
2RQ1
 
 | |
2RQ5
 
 | |
3TEE
 
 | | Crystal Structure of Salmonella FlgA in open form | | Descriptor: | CHLORIDE ION, Flagella basal body P-ring formation protein flgA, GLYCEROL | | Authors: | Matsunami, H, Samatey, F.A, Namba, K. | | Deposit date: | 2011-08-12 | | Release date: | 2012-08-15 | | Last modified: | 2016-07-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural flexibility of the periplasmic protein, FlgA, regulates flagellar P-ring assembly in Salmonella enterica
Sci Rep, 6, 2016
|
|
3VJP
 
 | |
7EHA
 
 | | Crystal structure of the flagellar hook cap from Salmonella enterica serovar Typhimurium | | Descriptor: | Basal-body rod modification protein FlgD | | Authors: | Matsunami, H, Yoon, Y.-H, Imada, K, Namba, K, Samatey, F.A. | | Deposit date: | 2021-03-29 | | Release date: | 2021-11-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of the bacterial flagellar hook cap provides insights into a hook assembly mechanism
Commun Biol, 4, 2021
|
|
7EH9
 
 | | Crystal structure of the flagellar hook cap fragment from Salmonella enterica serovar Typhimurium | | Descriptor: | Basal-body rod modification protein FlgD | | Authors: | Matsunami, H, Yoon, Y.-H, Imada, K, Namba, K, Samatey, F.A. | | Deposit date: | 2021-03-29 | | Release date: | 2021-11-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the bacterial flagellar hook cap provides insights into a hook assembly mechanism
Commun Biol, 4, 2021
|
|
3VKI
 
 | |
3W5Q
 
 | | Crystal structure of complexes of vitamin D receptor ligand binding domain with lithocholic acid derivatives | | Descriptor: | (5beta,9beta)-3-oxocholan-24-oic acid, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | Authors: | Masuno, H, Ikura, T, Ito, N. | | Deposit date: | 2013-02-05 | | Release date: | 2013-06-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of complexes of vitamin D receptor ligand-binding domain with lithocholic acid derivatives.
J.Lipid Res., 54, 2013
|
|
3W5R
 
 | | Crystal structure of complexes of vitamin D receptor ligand binding domain with lithocholic acid derivatives | | Descriptor: | (3beta,5beta,9beta)-3-(acetyloxy)cholan-24-oic acid, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | Authors: | Masuno, H, Ikura, T, Ito, N. | | Deposit date: | 2013-02-06 | | Release date: | 2013-06-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of complexes of vitamin D receptor ligand-binding domain with lithocholic acid derivatives.
J.Lipid Res., 54, 2013
|
|
3W5P
 
 | | Crystal structure of complexes of vitamin D receptor ligand binding domain with lithocholic acid derivatives | | Descriptor: | (3beta,5beta,14beta,17alpha)-3-hydroxycholan-24-oic acid, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | Authors: | Masuno, H, Ikura, T, Ito, N. | | Deposit date: | 2013-02-05 | | Release date: | 2013-06-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of complexes of vitamin D receptor ligand-binding domain with lithocholic acid derivatives.
J.Lipid Res., 54, 2013
|
|
3W5T
 
 | | Crystal structure of complexes of vitamin D receptor ligand binding domain with lithocholic acid derivatives | | Descriptor: | (3beta,5beta,9beta)-3-(propanoyloxy)cholan-24-oic acid, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | Authors: | Masuno, H, Ikura, T, Ito, N. | | Deposit date: | 2013-02-06 | | Release date: | 2013-06-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Crystal structures of complexes of vitamin D receptor ligand-binding domain with lithocholic acid derivatives.
J.Lipid Res., 54, 2013
|
|
2EJY
 
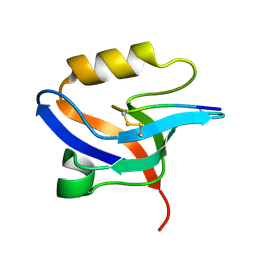 | |
2EV8
 
 | |
7X35
 
 | | Cryo-EM structure of Coxsackievirus B1 A-particle in complex with nAb 8A10 (CVB1-A:8A10) | | Descriptor: | 8A10 heavy chain, 8A10 light chain, VP2, ... | | Authors: | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | Deposit date: | 2022-02-28 | | Release date: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
4QY2
 
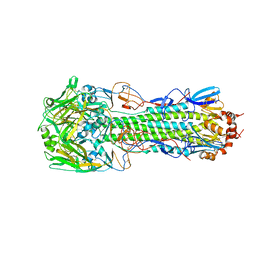 | | Structure of H10 from human-infecting H10N8 virus in complex with human receptor analog | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, N-acetyl-alpha-neuraminic acid, hemagglutinin | | Authors: | Wang, M, Zhang, W, Qi, J, Wang, F, Zhou, J, Bi, Y, Wu, Y, Sun, H, Liu, J, Huang, C, Li, X, Yan, J, Shu, Y, Shi, Y, Gao, G.F. | | Deposit date: | 2014-07-23 | | Release date: | 2015-01-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.399 Å) | | Cite: | Structural basis for preferential avian receptor binding by the human-infecting H10N8 avian influenza virus
Nat Commun, 6, 2015
|
|
4QY1
 
 | | Structure of H10 from human-infecting H10N8 in complex with avian receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, M, Zhang, W, Qi, J, Wang, F, Zhou, J, Bi, Y, Wu, Y, Sun, H, Liu, J, Huang, C, Li, X, Yan, J, Shu, Y, Shi, Y, Gao, G.F. | | Deposit date: | 2014-07-23 | | Release date: | 2015-01-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.594 Å) | | Cite: | Structural basis for preferential avian receptor binding by the human-infecting H10N8 avian influenza virus
Nat Commun, 6, 2015
|
|
6IO6
 
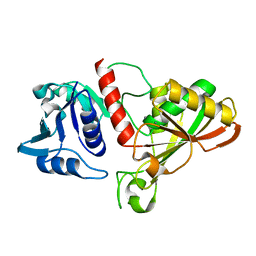 | |
