8IX3
 
 | | Cryo-EM structure of SARS-CoV-2 BA.4/5 spike protein in complex with 1G11 (local refinement) | | 分子名称: | BA.4/5 variant spike protein, heavy chain of 1G11, light chain of 1G11 | | 著者 | Sun, H, Jiang, Y, Zheng, Z, Zheng, Q, Li, S. | | 登録日 | 2023-03-31 | | 公開日 | 2023-11-15 | | 最終更新日 | 2023-12-13 | | 実験手法 | ELECTRON MICROSCOPY (3.98 Å) | | 主引用文献 | Structural basis for broad neutralization of human antibody against Omicron sublineages and evasion by XBB variant.
J.Virol., 97, 2023
|
|
7D6C
 
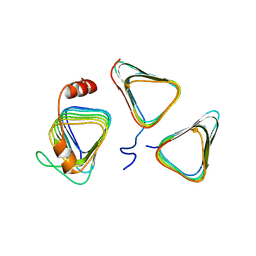 | | Crystal structure of CcmM N-terminal domain in complex with CcmN | | 分子名称: | Carbon dioxide concentrating mechanism protein CcmM, Carboxysome assembly protein CcmN | | 著者 | Sun, H, Cui, N, Han, S.J, Chen, Z.P, Xia, L.Y, Chen, Y, Jiang, Y.L, Zhou, C.Z. | | 登録日 | 2020-09-30 | | 公開日 | 2021-08-04 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.89 Å) | | 主引用文献 | Complex structure reveals CcmM and CcmN form a heterotrimeric adaptor in beta-carboxysome.
Protein Sci., 30, 2021
|
|
7XH0
 
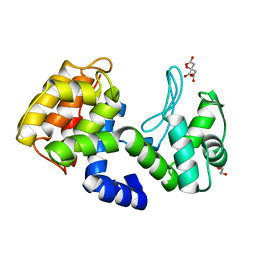 | | crystal structure of Csn-PD from Paenibacillus dendritiformis | | 分子名称: | 1,2-ETHANEDIOL, CITRATE ANION, Chitosanase | | 著者 | Sun, H.H, Cheng, Y.M, Cao, R, Liu, Q, Zhao, L. | | 登録日 | 2022-04-07 | | 公開日 | 2022-06-08 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.68 Å) | | 主引用文献 | crystal structure of Csn-PD from Paenibacillus dendritiformis
To Be Published
|
|
7X7U
 
 | | Cryo-EM structure of SARS-CoV-2 Delta variant spike protein in complex with three nAbs X01, X10 and X17 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, X01 heavy chain, ... | | 著者 | Sun, H, Liu, L, Zhang, T, Zheng, Q, Li, S, Xia, N. | | 登録日 | 2022-03-10 | | 公開日 | 2022-08-17 | | 最終更新日 | 2022-11-23 | | 実験手法 | ELECTRON MICROSCOPY (3.77 Å) | | 主引用文献 | The neutralizing breadth of antibodies targeting diverse conserved epitopes between SARS-CoV and SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7X7T
 
 | | Cryo-EM structure of SARS-CoV-2 spike protein in complex with three nAbs X01, X10 and X17 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, X01 heavy chain, ... | | 著者 | Sun, H, Liu, L, Zheng, Q, Li, S, Zhang, T, Xia, N. | | 登録日 | 2022-03-10 | | 公開日 | 2022-08-17 | | 最終更新日 | 2022-11-23 | | 実験手法 | ELECTRON MICROSCOPY (3.48 Å) | | 主引用文献 | The neutralizing breadth of antibodies targeting diverse conserved epitopes between SARS-CoV and SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7X7V
 
 | | Cryo-EM structure of SARS-CoV spike protein in complex with three nAbs X01, X10 and X17 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, X01 heavy chain, ... | | 著者 | Sun, H, Liu, L, Zhang, T, Zheng, Q, Li, S, Xia, N. | | 登録日 | 2022-03-10 | | 公開日 | 2022-08-17 | | 最終更新日 | 2022-11-23 | | 実験手法 | ELECTRON MICROSCOPY (3.83 Å) | | 主引用文献 | The neutralizing breadth of antibodies targeting diverse conserved epitopes between SARS-CoV and SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7XHH
 
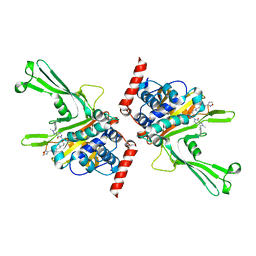 | | High-resolution X-ray cocrystal structure of USP7 in complex with X4 | | 分子名称: | 3-[4-(aminomethyl)phenyl]-6-[[1-[[2-chloranyl-4-(1,2,4-oxadiazol-3-yl)phenyl]methyl]-4-oxidanyl-piperidin-4-yl]methyl]-2-methyl-pyrazolo[4,3-d]pyrimidin-7-one, Ubiquitin carboxyl-terminal hydrolase 7 | | 著者 | Sun, H.B, Wen, X.A. | | 登録日 | 2022-04-08 | | 公開日 | 2023-04-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | High-resolution X-ray cocrystal structure of USP7 in complex with X4
To Be Published
|
|
7XHK
 
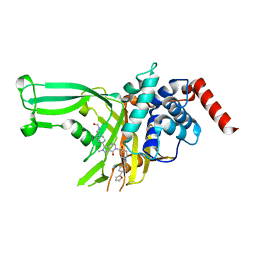 | | High-resolution X-ray cocrystal structure of USP7 in complex with LX04-46 | | 分子名称: | Ubiquitin carboxyl-terminal hydrolase 7, ~{N}-[[4-[6-[[1-[[2-chloranyl-4-(furan-2-yl)phenyl]methyl]-4-oxidanyl-piperidin-4-yl]methyl]-2-methyl-7-oxidanylidene-pyrazolo[4,3-d]pyrimidin-3-yl]phenyl]methyl]methanamide | | 著者 | Sun, H.B, Wen, X.A. | | 登録日 | 2022-04-08 | | 公開日 | 2023-06-14 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | High-resolution X-ray cocrystal structure of USP7 in complex with LX04-46
To Be Published
|
|
6K5O
 
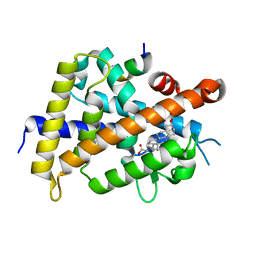 | | Development of Novel Lithocholic Acid Derivatives as Vitamin D Receptor Agonists | | 分子名称: | (4~{R})-4-[(3~{R},5~{R},8~{R},9~{S},10~{S},13~{R},14~{S},17~{R})-10,13-dimethyl-3-methylsulfonyloxy-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1~{H}-cyclopenta[a]phenanthren-17-yl]pentanoic acid, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | 著者 | Masuno, H, Kagechika, H, Ito, N. | | 登録日 | 2019-05-29 | | 公開日 | 2019-07-24 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Development of novel lithocholic acid derivatives as vitamin D receptor agonists.
Bioorg.Med.Chem., 27, 2019
|
|
7C7W
 
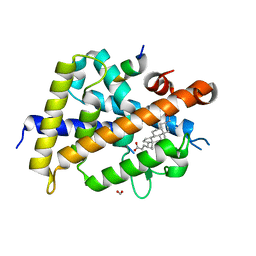 | | Vitamin D3 receptor/lithochoric acid derivative complex | | 分子名称: | (4R)-4-[(3S,5R,8R,9S,10S,13R,14S,17R)-10,13-dimethyl-3-(2-methyl-2-oxidanyl-propyl)-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthren-17-yl]pentanoic acid, FORMIC ACID, Mediator of RNA polymerase II transcription subunit 1, ... | | 著者 | Masuno, H, Numoto, N, Kagechika, H, Ito, N. | | 登録日 | 2020-05-26 | | 公開日 | 2021-01-20 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Lithocholic Acid Derivatives as Potent Vitamin D Receptor Agonists.
J.Med.Chem., 64, 2021
|
|
5JXL
 
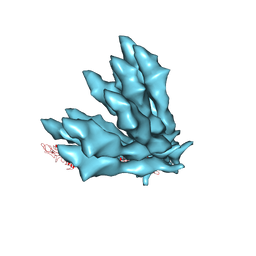 | |
1A13
 
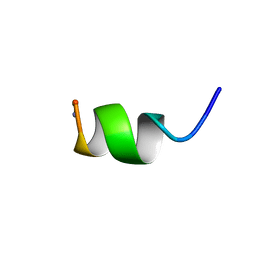 | | G PROTEIN-BOUND CONFORMATION OF MASTOPARAN-X, NMR, 14 STRUCTURES | | 分子名称: | MASTOPARAN-X | | 著者 | Kusunoki, H, Wakamatsu, K, Sato, K, Miyazawa, T, Kohno, T. | | 登録日 | 1997-12-20 | | 公開日 | 1999-01-13 | | 最終更新日 | 2022-02-16 | | 実験手法 | SOLUTION NMR | | 主引用文献 | G protein-bound conformation of mastoparan-X: heteronuclear multidimensional transferred nuclear overhauser effect analysis of peptide uniformly enriched with 13C and 15N.
Biochemistry, 37, 1998
|
|
7C7V
 
 | | Vitamin D3 receptor/lithochoric acid derivative complex | | 分子名称: | (4R)-4-[(3R,5R,8R,9S,10S,13R,14S,17R)-10,13-dimethyl-3-(2-methyl-2-oxidanyl-propyl)-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthren-17-yl]pentanoic acid, FORMIC ACID, Mediator of RNA polymerase II transcription subunit 1, ... | | 著者 | Masuno, H, Numoto, N, Kagechika, H, Ito, N. | | 登録日 | 2020-05-26 | | 公開日 | 2021-01-20 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Lithocholic Acid Derivatives as Potent Vitamin D Receptor Agonists.
J.Med.Chem., 64, 2021
|
|
1UI7
 
 | | Site-directed mutagenesis of His433 involved in binding of copper ion in Arthrobacter globiformis amine oxidase | | 分子名称: | COPPER (II) ION, Phenylethylamine oxidase | | 著者 | Matsunami, H, Okajima, T, Hirota, S, Yamaguchi, H, Hori, H, Kuroda, S, Tanizawa, K. | | 登録日 | 2003-07-15 | | 公開日 | 2004-04-20 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Chemical rescue of a site-specific mutant of bacterial copper amine oxidase for generation of the topa quinone cofactor
Biochemistry, 43, 2004
|
|
1UI8
 
 | | Site-directed mutagenesis of His592 involved in binding of copper ion in Arthrobacter globiformis amine oxidase | | 分子名称: | COPPER (II) ION, Phenylethylamine oxidase | | 著者 | Matsunami, H, Okajima, T, Hirota, S, Yamaguchi, H, Hori, H, Kuroda, S, Tanizawa, K. | | 登録日 | 2003-07-15 | | 公開日 | 2004-04-20 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Chemical rescue of a site-specific mutant of bacterial copper amine oxidase for generation of the topa quinone cofactor
Biochemistry, 43, 2004
|
|
3VKI
 
 | |
2UYG
 
 | |
1K1V
 
 | | Solution Structure of the DNA-Binding Domain of MafG | | 分子名称: | MafG | | 著者 | Kusunoki, H, Motohashi, H, Katsuoka, F, Morohashi, A, Yamamoto, M, Tanaka, T. | | 登録日 | 2001-09-25 | | 公開日 | 2002-04-10 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the DNA-binding domain of MafG.
Nat.Struct.Biol., 9, 2002
|
|
3VJP
 
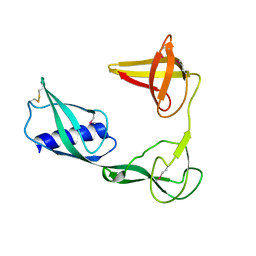 | |
3HJR
 
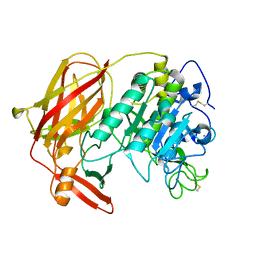 | | Crystal structure of serine protease of Aeromonas sobria | | 分子名称: | CALCIUM ION, Extracellular serine protease | | 著者 | Utsunomiya, H, Tsuge, H, Kobayashi, H, Okamoto, K. | | 登録日 | 2009-05-22 | | 公開日 | 2009-06-02 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Structural basis for the kexin-like serine protease from Aeromonas sobria as a sepsis-causing factor
J.Biol.Chem., 284, 2009
|
|
2RQ1
 
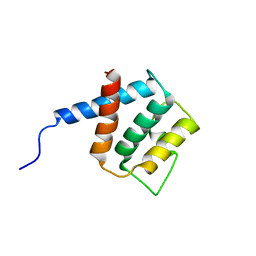 | |
2RQ5
 
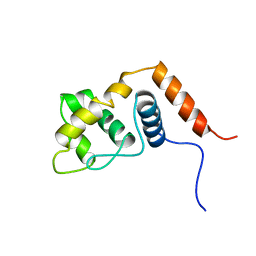 | |
3W5P
 
 | | Crystal structure of complexes of vitamin D receptor ligand binding domain with lithocholic acid derivatives | | 分子名称: | (3beta,5beta,14beta,17alpha)-3-hydroxycholan-24-oic acid, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | 著者 | Masuno, H, Ikura, T, Ito, N. | | 登録日 | 2013-02-05 | | 公開日 | 2013-06-26 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structures of complexes of vitamin D receptor ligand-binding domain with lithocholic acid derivatives.
J.Lipid Res., 54, 2013
|
|
3W5T
 
 | | Crystal structure of complexes of vitamin D receptor ligand binding domain with lithocholic acid derivatives | | 分子名称: | (3beta,5beta,9beta)-3-(propanoyloxy)cholan-24-oic acid, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | 著者 | Masuno, H, Ikura, T, Ito, N. | | 登録日 | 2013-02-06 | | 公開日 | 2013-06-26 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.29 Å) | | 主引用文献 | Crystal structures of complexes of vitamin D receptor ligand-binding domain with lithocholic acid derivatives.
J.Lipid Res., 54, 2013
|
|
3W5Q
 
 | | Crystal structure of complexes of vitamin D receptor ligand binding domain with lithocholic acid derivatives | | 分子名称: | (5beta,9beta)-3-oxocholan-24-oic acid, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | 著者 | Masuno, H, Ikura, T, Ito, N. | | 登録日 | 2013-02-05 | | 公開日 | 2013-06-26 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structures of complexes of vitamin D receptor ligand-binding domain with lithocholic acid derivatives.
J.Lipid Res., 54, 2013
|
|
