1CNS
 
 | |
6JN8
 
 | | Structure of H216A mutant open form peptidoglycan peptidase | | Descriptor: | Peptidase M23, SULFATE ION, ZINC ION | | Authors: | Min, K.J, An, D.R, Yoon, H.J, Suh, S.W, Lee, H.H. | | Deposit date: | 2019-03-13 | | Release date: | 2020-01-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.106 Å) | | Cite: | Peptidoglycan reshaping by a noncanonical peptidase for helical cell shape in Campylobacter jejuni.
Nat Commun, 11, 2020
|
|
6JMX
 
 | | Structure of open form of peptidoglycan peptidase | | Descriptor: | D(-)-TARTARIC ACID, GLYCEROL, Peptidase M23, ... | | Authors: | Min, K.J, An, D.R, Yoon, H.J, Suh, S.W, Lee, H.H. | | Deposit date: | 2019-03-13 | | Release date: | 2020-01-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.859 Å) | | Cite: | Peptidoglycan reshaping by a noncanonical peptidase for helical cell shape in Campylobacter jejuni.
Nat Commun, 11, 2020
|
|
6JN1
 
 | | Structure of H247A mutant peptidoglycan peptidase complex with penta peptide | | Descriptor: | C0O-DAL-DAL, Peptidase M23, ZINC ION | | Authors: | Min, K.J, An, D.R, Yoon, H.J, Suh, S.W, Lee, H.H. | | Deposit date: | 2019-03-13 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.382 Å) | | Cite: | Peptidoglycan reshaping by a noncanonical peptidase for helical cell shape in Campylobacter jejuni.
Nat Commun, 11, 2020
|
|
6JMZ
 
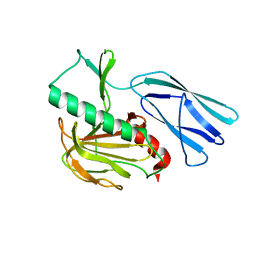 | | Structure of H247A mutant open form peptidoglycan peptidase | | Descriptor: | Peptidase M23, ZINC ION | | Authors: | Min, K.J, An, D.R, Yoon, H.J, Suh, S.W, Lee, H.H. | | Deposit date: | 2019-03-13 | | Release date: | 2020-01-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Peptidoglycan reshaping by a noncanonical peptidase for helical cell shape in Campylobacter jejuni.
Nat Commun, 11, 2020
|
|
6JN0
 
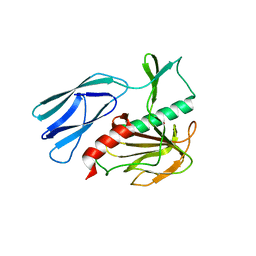 | | Structure of H247A mutant peptidoglycan peptidase complex with tetra-tri peptide | | Descriptor: | C0O-DAL-API, Peptidase M23, ZINC ION | | Authors: | Min, K.J, An, D.R, Yoon, H.J, Suh, S.W, Lee, H.H. | | Deposit date: | 2019-03-13 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.164 Å) | | Cite: | Peptidoglycan reshaping by a noncanonical peptidase for helical cell shape in Campylobacter jejuni.
Nat Commun, 11, 2020
|
|
6JN7
 
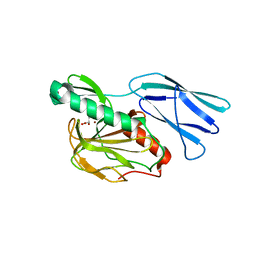 | | Structure of H216A mutant closed form peptidoglycan peptidase | | Descriptor: | D(-)-TARTARIC ACID, Peptidase M23, ZINC ION | | Authors: | Min, K.J, An, D.R, Yoon, H.J, Suh, S.W, Lee, H.H. | | Deposit date: | 2019-03-13 | | Release date: | 2020-01-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Peptidoglycan reshaping by a noncanonical peptidase for helical cell shape in Campylobacter jejuni.
Nat Commun, 11, 2020
|
|
6JMY
 
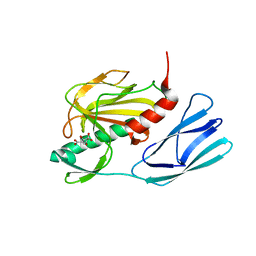 | | Structure of wild type closed form of peptidoglycan peptidase | | Descriptor: | CITRIC ACID, Peptidase M23, ZINC ION | | Authors: | Min, K.J, An, D.R, Yoon, H.J, Suh, S.W, Lee, H.H. | | Deposit date: | 2019-03-13 | | Release date: | 2020-01-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.661 Å) | | Cite: | Peptidoglycan reshaping by a noncanonical peptidase for helical cell shape in Campylobacter jejuni.
Nat Commun, 11, 2020
|
|
6KV1
 
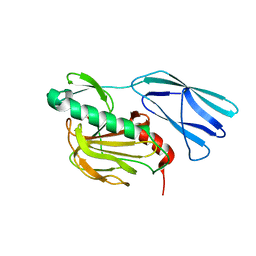 | | Structure of wild type closed form of peptidoglycan peptidase ZN SAD | | Descriptor: | CITRIC ACID, Peptidase M23, ZINC ION | | Authors: | Min, K.J, An, D.R, Yoon, H.J, Suh, S.W, Lee, H.H. | | Deposit date: | 2019-09-03 | | Release date: | 2020-01-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.722 Å) | | Cite: | Peptidoglycan reshaping by a noncanonical peptidase for helical cell shape in Campylobacter jejuni.
Nat Commun, 11, 2020
|
|
1HYE
 
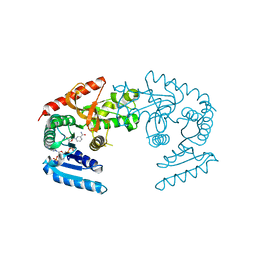 | | CRYSTAL STRUCTURE OF THE MJ0490 GENE PRODUCT, THE FAMILY OF LACTATE/MALATE DEHYDROGENASE, DIMERIC STRUCTURE | | Descriptor: | L-LACTATE/MALATE DEHYDROGENASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Lee, B.I, Chang, C, Cho, S.-J, Suh, S.W. | | Deposit date: | 2001-01-19 | | Release date: | 2001-04-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the MJ0490 gene product of the hyperthermophilic archaebacterium Methanococcus jannaschii, a novel member of the lactate/malate family of dehydrogenases.
J.Mol.Biol., 307, 2001
|
|
1HYG
 
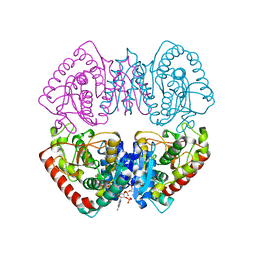 | | Crystal structure of MJ0490 gene product, the family of lactate/malate dehydrogenase | | Descriptor: | L-LACTATE/MALATE DEHYDROGENASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Lee, B.I, Chang, C, Cho, S.-J, Suh, S.W. | | Deposit date: | 2001-01-19 | | Release date: | 2001-04-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the MJ0490 gene product of the hyperthermophilic archaebacterium Methanococcus jannaschii, a novel member of the lactate/malate family of dehydrogenases
J.Mol.Biol., 307, 2001
|
|
1J2Y
 
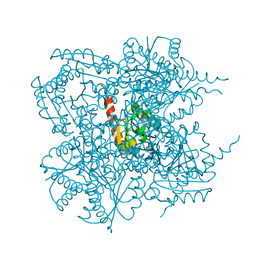 | | Crystal structure of the type II 3-dehydroquinase | | Descriptor: | 1,3,4-TRIHYDROXY-5-OXO-CYCLOHEXANECARBOXYLIC ACID, 3-dehydroquinate dehydratase | | Authors: | Lee, B.I, Kwak, J.E, Suh, S.W. | | Deposit date: | 2003-01-15 | | Release date: | 2003-06-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the type II 3-dehydroquinase from Helicobacter pylori
Proteins, 51, 2003
|
|
1IX1
 
 | | Crystal Structure of P.aeruginosa Peptide deformylase Complexed with Antibiotic Actinonin | | Descriptor: | (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, ACTINONIN, ZINC ION, ... | | Authors: | Kim, H.-W, Yoon, H.-J, Lee, J.Y, Han, B.W, Yang, J.K, Lee, B.I, Ahn, H.J, Lee, H.H, Suh, S.W. | | Deposit date: | 2002-06-07 | | Release date: | 2003-09-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of peptide deformylase from Staphylococcus aureus in complex with actinonin, a naturally occurring antibacterial agent
Proteins, 57, 2004
|
|
1J2Z
 
 | | Crystal structure of UDP-N-acetylglucosamine acyltransferase | | Descriptor: | Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase, L(+)-TARTARIC ACID, SULFATE ION, ... | | Authors: | Lee, B.I, Suh, S.W. | | Deposit date: | 2003-01-15 | | Release date: | 2004-01-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of UDP-N-acetylglucosamine acyltransferase from Helicobacter pylori
Proteins, 53, 2003
|
|
4FYB
 
 | | Structural and functional characterizations of a thioredoxin-fold protein from Helicobacter pylori | | Descriptor: | GLYCEROL, Thiol:disulfide interchange protein (DsbC) | | Authors: | Yoon, J.Y, Kim, J, Lee, S.J, Im, H.N, Kim, H.S, Yoon, H, An, D.R, Kim, J.Y, Kim, S, Han, B.W, Suh, S.W. | | Deposit date: | 2012-07-04 | | Release date: | 2013-05-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and functional characterization of HP0377, a thioredoxin-fold protein from Helicobacter pylori
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4FBB
 
 | | Structure of mutant RIP from barley seeds in complex with adenine (AMP-incubated) | | Descriptor: | ADENINE, Protein synthesis inhibitor I | | Authors: | Lee, B.-G, Kim, M.K, Suh, S.W, Song, H.K. | | Deposit date: | 2012-05-22 | | Release date: | 2012-10-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of the ribosome-inactivating protein from barley seeds reveal a unique activation mechanism.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
1JUV
 
 | |
4FBH
 
 | | Structure of RIP from barley seeds | | Descriptor: | ADENOSINE MONOPHOSPHATE, Protein synthesis inhibitor I | | Authors: | Lee, B.-G, Kim, M.K, Suh, S.W, Song, H.K. | | Deposit date: | 2012-05-23 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of the ribosome-inactivating protein from barley seeds reveal a unique activation mechanism.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4FIR
 
 | | Crystal structure of pyridoxal biosynthesis lyase PdxS from Pyrococcus | | Descriptor: | Pyridoxal biosynthesis lyase pdxS, RIBOSE-5-PHOSPHATE | | Authors: | Matsuura, A, Yoon, J.Y, Yoon, H.J, Lee, H.H, Suh, S.W. | | Deposit date: | 2012-06-11 | | Release date: | 2012-11-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of pyridoxal biosynthesis lyase PdxS from Pyrococcus horikoshii.
Mol.Cells, 34, 2012
|
|
4FBC
 
 | | Structure of mutant RIP from barley seeds in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Protein synthesis inhibitor I | | Authors: | Lee, B.-G, Kim, M.K, Suh, S.W, Song, H.K. | | Deposit date: | 2012-05-22 | | Release date: | 2012-10-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of the ribosome-inactivating protein from barley seeds reveal a unique activation mechanism.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4GSQ
 
 | | Structural basis for the inhibition of Mycobacterium tuberculosis L,D-transpeptidase by meropenem, a drug effective against extensively drug-resistant strains | | Descriptor: | CALCIUM ION, GLYCEROL, Probable conserved lipoprotein LPPS | | Authors: | Kim, H.S, Kim, J, Im, H.N, Yoon, J.Y, An, D.R, Yoon, H.J, Kim, J.Y, Min, H.K, Kim, S.-J, Lee, J.Y, Han, B.W, Suh, S.W. | | Deposit date: | 2012-08-28 | | Release date: | 2013-02-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the inhibition of Mycobacterium tuberculosis L,D-transpeptidase by meropenem, a drug effective against extensively drug-resistant strains
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4GSU
 
 | | Structural basis for the inhibition of Mycobacterium tuberculosis L,D-transpeptidase by meropenem, a drug effective against extensively drug-resistant strains | | Descriptor: | (2S,3R,4S)-4-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-2-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-3-methyl-3,4-dihydro-2H-pyrrole-5-carboxylic acid, Probable conserved lipoprotein LPPS | | Authors: | Kim, H.S, Kim, J, Im, H.N, Yoon, J.Y, An, D.R, Yoon, H.J, Kim, J.Y, Min, H.K, Kim, S.-J, Lee, J.Y, Han, B.W, Suh, S.W. | | Deposit date: | 2012-08-28 | | Release date: | 2013-02-27 | | Last modified: | 2022-02-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the inhibition of Mycobacterium tuberculosis L,D-transpeptidase by meropenem, a drug effective against extensively drug-resistant strains
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4FIQ
 
 | | Crystal structure of pyridoxal biosynthesis lyase PdxS from Pyrococcus horikoshii | | Descriptor: | Pyridoxal biosynthesis lyase pdxS | | Authors: | Matsuura, A, Yoon, J.Y, Yoon, H.J, Lee, H.H, Suh, S.W. | | Deposit date: | 2012-06-11 | | Release date: | 2012-11-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of pyridoxal biosynthesis lyase PdxS from Pyrococcus horikoshii.
Mol.Cells, 34, 2012
|
|
4FB9
 
 | | Structure of mutant RIP from barley seeds | | Descriptor: | Protein synthesis inhibitor I | | Authors: | Lee, B.-G, Kim, M.K, Suh, S.W, Song, H.K. | | Deposit date: | 2012-05-22 | | Release date: | 2012-10-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structures of the ribosome-inactivating protein from barley seeds reveal a unique activation mechanism.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
1KCT
 
 | | ALPHA1-ANTITRYPSIN | | Descriptor: | ALPHA1-ANTITRYPSIN | | Authors: | Song, H.K, Suh, S.W. | | Deposit date: | 1996-08-06 | | Release date: | 1997-01-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.46 Å) | | Cite: | Crystal structure of an uncleaved alpha 1-antitrypsin reveals the conformation of its inhibitory reactive loop.
FEBS Lett., 377, 1995
|
|
