7K8B
 
 | |
3OW7
 
 | |
6WTI
 
 | | The Cryo-EM structure of the ubiquinol oxidase from Escherichia coli | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, COPPER (II) ION, Cytochrome o ubiquinol oxidase, ... | | Authors: | Su, C.-C. | | Deposit date: | 2020-05-02 | | Release date: | 2021-01-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.38 Å) | | Cite: | A 'Build and Retrieve' methodology to simultaneously solve cryo-EM structures of membrane proteins.
Nat.Methods, 18, 2021
|
|
3NE5
 
 | |
8EKY
 
 | | Cryo-EM structure of the human PRDX4-ErP46 complex | | Descriptor: | Peroxiredoxin-4, Thioredoxin domain-containing protein 5 | | Authors: | Su, C.C. | | Deposit date: | 2022-09-22 | | Release date: | 2023-05-03 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | High-resolution structural-omics of human liver enzymes.
Cell Rep, 42, 2023
|
|
8EKW
 
 | | Cryo-EM structure of human PRDX4 | | Descriptor: | Peroxiredoxin-4 | | Authors: | Su, C.C. | | Deposit date: | 2022-09-22 | | Release date: | 2023-05-03 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | High-resolution structural-omics of human liver enzymes.
Cell Rep, 42, 2023
|
|
8EM2
 
 | |
3OOC
 
 | | Crystal structure of the membrane fusion protein CusB from Escherichia coli | | Descriptor: | Cation efflux system protein cusB | | Authors: | Su, C.-C, Yang, F, Long, F, Reyon, D, Routh, M.D, Kuo, D.W, Mokhtari, A.K, Van Ornam, J.D, Rabe, K.L, Hoy, J.A, Lee, Y.J, Rajashankar, K.R, Yu, E.W. | | Deposit date: | 2010-08-30 | | Release date: | 2010-12-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.404 Å) | | Cite: | Crystal structure of the membrane fusion protein CusB from Escherichia coli.
J.Mol.Biol., 393, 2009
|
|
3OPO
 
 | |
4K34
 
 | |
4MT4
 
 | |
4K7K
 
 | |
4K7R
 
 | | Crystal structures of CusC review conformational changes accompanying folding and transmembrane channel formation | | Descriptor: | (2S)-1-(pentanoyloxy)propan-2-yl hexanoate, Cation efflux system protein CusC | | Authors: | Su, C.-C, Lei, H.-T, Bolla, J.R, Yu, E.W. | | Deposit date: | 2013-04-17 | | Release date: | 2013-10-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.094 Å) | | Cite: | Crystal Structures of CusC Review Conformational Changes Accompanying Folding and Transmembrane Channel Formation.
J.Mol.Biol., 426, 2014
|
|
4NB5
 
 | |
9BFT
 
 | | Cryo-EM co-structure of AcrB with CU244 | | Descriptor: | (2S)-1-{[(1R,5R)-3-azabicyclo[3.1.0]hexan-6-yl]amino}-3-(3,5-dichlorophenoxy)propan-2-ol, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Multidrug efflux pump subunit AcrB | | Authors: | Su, C.C. | | Deposit date: | 2024-04-18 | | Release date: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.44 Å) | | Cite: | Bacterial efflux pump modulators prevent bacterial growth in macrophages and under broth conditions that mimic the host environment.
mBio, 14, 2023
|
|
9BFN
 
 | | Cryo-EM co-structure of AcrB with the CU232 efflux pump inhibitor | | Descriptor: | (2R)-1-(4-aminopiperidin-1-yl)-3-[3-(trifluoromethyl)phenoxy]propan-2-ol, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Multidrug efflux pump subunit AcrB | | Authors: | Su, C.C. | | Deposit date: | 2024-04-18 | | Release date: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | Bacterial efflux pump modulators prevent bacterial growth in macrophages and under broth conditions that mimic the host environment.
mBio, 14, 2023
|
|
9BFH
 
 | | Cryo-EM co-structure of AcrB with the CU032 efflux pump inhibitor | | Descriptor: | (2S)-1-[(3R)-3-aminopyrrolidin-1-yl]-3-(3,4-dichlorophenoxy)propan-2-ol, Multidrug efflux pump subunit AcrB | | Authors: | Su, C.C. | | Deposit date: | 2024-04-17 | | Release date: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.62 Å) | | Cite: | Bacterial efflux pump modulators prevent bacterial growth in macrophages and under broth conditions that mimic the host environment.
mBio, 14, 2023
|
|
9BFM
 
 | | Cryo-EM co-structure of AcrB with the EPM35 efflux pump inhibitor | | Descriptor: | (2S)-1-(3,4-dichlorophenoxy)-3-(4-{[4-(trifluoromethyl)pyrimidin-2-yl]amino}piperidin-1-yl)propan-2-ol, Multidrug efflux pump subunit AcrB | | Authors: | Su, C.C. | | Deposit date: | 2024-04-18 | | Release date: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | Bacterial efflux pump modulators prevent bacterial growth in macrophages and under broth conditions that mimic the host environment.
mBio, 14, 2023
|
|
6N40
 
 | |
4R0C
 
 | | Crystal structure of the Alcanivorax borkumensis YdaH transporter reveals an unusual topology | | Descriptor: | AbgT putative transporter family, DODECYL-BETA-D-MALTOSIDE, SODIUM ION, ... | | Authors: | Su, C.-C, Bolla, J.R, Yu, E.W. | | Deposit date: | 2014-07-30 | | Release date: | 2015-04-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.963 Å) | | Cite: | Crystal structure of the Alcanivorax borkumensis YdaH transporter reveals an unusual topology.
Nat Commun, 6, 2015
|
|
4DNR
 
 | |
3H94
 
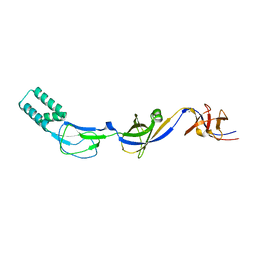 | | Crystal structure of the membrane fusion protein CusB from Escherichia coli | | Descriptor: | Cation efflux system protein cusB, SILVER ION | | Authors: | Su, C.-C, Yang, F, Long, F, Reyon, D, Routh, M.D, Kuo, D.W, Mokhtari, A.K, Van Ornam, J.D, Rabe, K.L, Hoy, J.A, Lee, Y.J, Rajashankar, K.R, Yu, E.W. | | Deposit date: | 2009-04-30 | | Release date: | 2009-08-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.84 Å) | | Cite: | Crystal structure of the membrane fusion protein CusB from Escherichia coli
J.Mol.Biol., 393, 2009
|
|
4R1I
 
 | |
4MT1
 
 | |
4MT0
 
 | |
