7K8D
 
 | |
7N6B
 
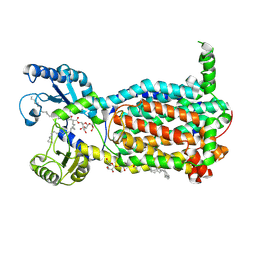 | | Structure of MmpL3 reconstituted into lipid nanodisc in the TMM bound state | | Descriptor: | 6-O-[(2S)-2-{(1S)-18-[(1R,2R)-2-hexylcyclopropyl]-1-hydroxyoctadecyl}tricosanoyl]-alpha-D-glucopyranosyl alpha-D-glucopyranoside, MmpL3 transporter | | Authors: | Su, C.C, Yu, E. | | Deposit date: | 2021-06-08 | | Release date: | 2021-09-01 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Structures of the mycobacterial membrane protein MmpL3 reveal its mechanism of lipid transport.
Plos Biol., 19, 2021
|
|
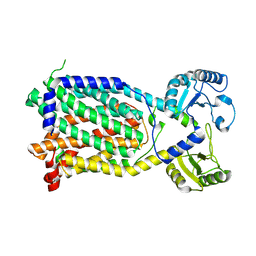
7K8C
 
 | |
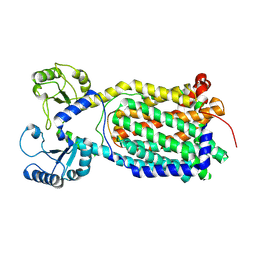
7K8A
 
 | |
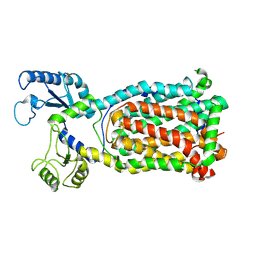
7K8B
 
 | |
6N40
 
 | |
7K7M
 
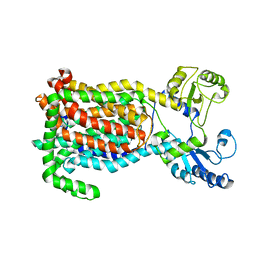 | | Crystal Structure of a membrane protein | | Descriptor: | Drug exporters of the RND superfamily-like protein, alpha-D-glucopyranose-(1-1)-6-O-decanoyl-alpha-D-glucopyranose | | Authors: | Su, C.-C. | | Deposit date: | 2020-09-23 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.33 Å) | | Cite: | Structures of the mycobacterial membrane protein MmpL3 reveal its mechanism of lipid transport.
Plos Biol., 19, 2021
|
|
8WPY
 
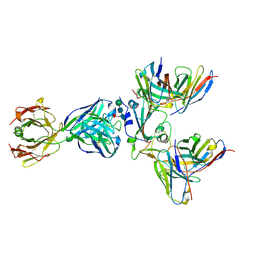 | | Cryo-EM structure of SARS-CoV-2 receptor-binding domain (RBD) complexed with CB6 mutant,S309, and S304 antibodies | | Descriptor: | CB6 fab mutant heavy chain, CB6 fab mutant light chain, S304 fab heavy chain, ... | | Authors: | Su, C, Qi, J.X, Gao, G.F. | | Deposit date: | 2023-10-10 | | Release date: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | An improved design method enables the ineffective etesevimab broadly and efficiently against SARS-CoV-2 Omicron subvariants
To Be Published
|
|
8WPW
 
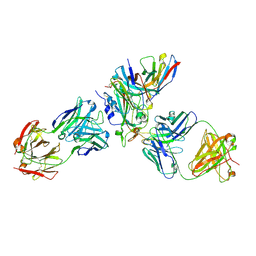 | | Cryo-EM structure of SARS-CoV-2 XBB.1.5 receptor-binding domain (RBD) complexed with CB6 mutant,S309, and S304 antibodies | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CB6 fab mutant heavy chain, CB6 fab mutant light chain, ... | | Authors: | Su, C, Qi, J.X, Gao, G.F. | | Deposit date: | 2023-10-10 | | Release date: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | An improved design method enables the ineffective etesevimab broadly and efficiently against SARS-CoV-2 Omicron subvariants
To Be Published
|
|
7JZH
 
 | |
8EM2
 
 | |
4K7K
 
 | |
7JZ6
 
 | |
6WTI
 
 | | The Cryo-EM structure of the ubiquinol oxidase from Escherichia coli | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, COPPER (II) ION, Cytochrome o ubiquinol oxidase, ... | | Authors: | Su, C.-C. | | Deposit date: | 2020-05-02 | | Release date: | 2021-01-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.38 Å) | | Cite: | A 'Build and Retrieve' methodology to simultaneously solve cryo-EM structures of membrane proteins.
Nat.Methods, 18, 2021
|
|
4K7R
 
 | | Crystal structures of CusC review conformational changes accompanying folding and transmembrane channel formation | | Descriptor: | (2S)-1-(pentanoyloxy)propan-2-yl hexanoate, Cation efflux system protein CusC | | Authors: | Su, C.-C, Lei, H.-T, Bolla, J.R, Yu, E.W. | | Deposit date: | 2013-04-17 | | Release date: | 2013-10-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.094 Å) | | Cite: | Crystal Structures of CusC Review Conformational Changes Accompanying Folding and Transmembrane Channel Formation.
J.Mol.Biol., 426, 2014
|
|
3K07
 
 | | Crystal structure of CusA | | Descriptor: | Cation efflux system protein cusA | | Authors: | Su, C.-C. | | Deposit date: | 2009-09-24 | | Release date: | 2010-09-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.521 Å) | | Cite: | Crystal structures of the CusA efflux pump suggest methionine-mediated metal transport.
Nature, 467, 2010
|
|
3KSO
 
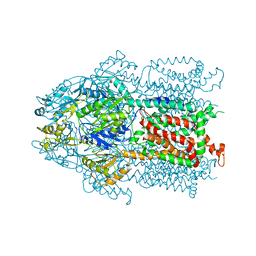 | | Structure and Mechanism of the Heavy Metal Transporter CusA | | Descriptor: | Cation efflux system protein cusA, SILVER ION | | Authors: | Su, C.-C. | | Deposit date: | 2009-11-23 | | Release date: | 2010-09-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (4.367 Å) | | Cite: | Crystal structures of the CusA efflux pump suggest methionine-mediated metal transport.
Nature, 467, 2010
|
|
3KSS
 
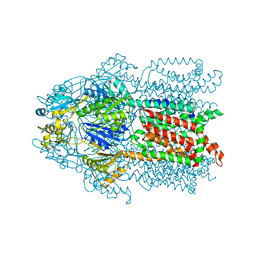 | | Structure and Mechanism of the Heavy Metal Transporter CusA | | Descriptor: | COPPER (I) ION, Cation efflux system protein cusA | | Authors: | Su, C.-C. | | Deposit date: | 2009-11-23 | | Release date: | 2010-09-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.88 Å) | | Cite: | Crystal structures of the CusA efflux pump suggest methionine-mediated metal transport.
Nature, 467, 2010
|
|
2HQF
 
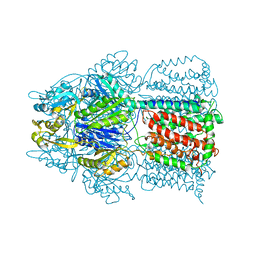 | | Conformation of the AcrB Multidrug Efflux Pump in Mutants of the Putative Proton Relay Pathway | | Descriptor: | Acriflavine resistance protein B | | Authors: | Su, C.-C, Li, M, Gu, R, Takatsuka, Y, McDermott, G, Nikaido, H, Yu, E.W. | | Deposit date: | 2006-07-18 | | Release date: | 2007-04-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.38 Å) | | Cite: | Conformation of the AcrB multidrug efflux pump in mutants of the putative proton relay pathway
J.Bacteriol., 188, 2006
|
|
8EKY
 
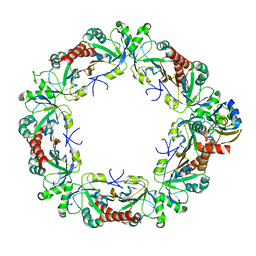 | | Cryo-EM structure of the human PRDX4-ErP46 complex | | Descriptor: | Peroxiredoxin-4, Thioredoxin domain-containing protein 5 | | Authors: | Su, C.C. | | Deposit date: | 2022-09-22 | | Release date: | 2023-05-03 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | High-resolution structural-omics of human liver enzymes.
Cell Rep, 42, 2023
|
|
8EKW
 
 | | Cryo-EM structure of human PRDX4 | | Descriptor: | Peroxiredoxin-4 | | Authors: | Su, C.C. | | Deposit date: | 2022-09-22 | | Release date: | 2023-05-03 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | High-resolution structural-omics of human liver enzymes.
Cell Rep, 42, 2023
|
|
4K34
 
 | |
3T53
 
 | | Crystal structures of the extrusion state of the CusBA adaptor-transporter complex | | Descriptor: | COPPER (II) ION, Cation efflux system protein CusA, Cation efflux system protein CusB | | Authors: | Su, C.-C, Long, F, Yu, E.W. | | Deposit date: | 2011-07-26 | | Release date: | 2012-06-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.37 Å) | | Cite: | Charged Amino Acids (R83, E567, D617, E625, R669, and K678) of CusA Are Required for Metal Ion Transport in the Cus Efflux System.
J.Mol.Biol., 422, 2012
|
|
6VQQ
 
 | |
7W8S
 
 | | Structure of SARS-CoV-2 spike receptor-binding domain Y453F mutation complexed with American mink ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S1, ... | | Authors: | Su, C, Qi, J.X, Gao, G.F. | | Deposit date: | 2021-12-08 | | Release date: | 2022-08-17 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Molecular Basis of Mink ACE2 Binding to SARS-CoV-2 and Its Mink-Derived Variants.
J.Virol., 96, 2022
|
|
