6OWS
 
 | |
3H94
 
 | | Crystal structure of the membrane fusion protein CusB from Escherichia coli | | Descriptor: | Cation efflux system protein cusB, SILVER ION | | Authors: | Su, C.-C, Yang, F, Long, F, Reyon, D, Routh, M.D, Kuo, D.W, Mokhtari, A.K, Van Ornam, J.D, Rabe, K.L, Hoy, J.A, Lee, Y.J, Rajashankar, K.R, Yu, E.W. | | Deposit date: | 2009-04-30 | | Release date: | 2009-08-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.84 Å) | | Cite: | Crystal structure of the membrane fusion protein CusB from Escherichia coli
J.Mol.Biol., 393, 2009
|
|
3KSS
 
 | | Structure and Mechanism of the Heavy Metal Transporter CusA | | Descriptor: | COPPER (I) ION, Cation efflux system protein cusA | | Authors: | Su, C.-C. | | Deposit date: | 2009-11-23 | | Release date: | 2010-09-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.88 Å) | | Cite: | Crystal structures of the CusA efflux pump suggest methionine-mediated metal transport.
Nature, 467, 2010
|
|
5T0O
 
 | | Crystal Structure of a membrane protein | | Descriptor: | CmeB | | Authors: | Su, C.-C, Yu, E.W. | | Deposit date: | 2016-08-16 | | Release date: | 2017-09-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structures and transport dynamics of a Campylobacter jejuni multidrug efflux pump.
Nat Commun, 8, 2017
|
|
5KHS
 
 | |
5KHN
 
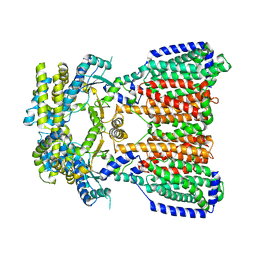 | |
8T3U
 
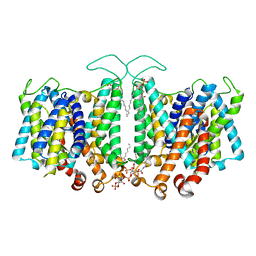 | | Cryo-EM Analysis of AE1 Structure in 100 mM NaCl Buffer: Form2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Band 3 anion transport protein, DIUNDECYL PHOSPHATIDYL CHOLINE, ... | | Authors: | Su, C.C. | | Deposit date: | 2023-06-07 | | Release date: | 2024-05-22 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Cryo-EM structure of the band 3 anion transport protein
To Be Published
|
|
8T45
 
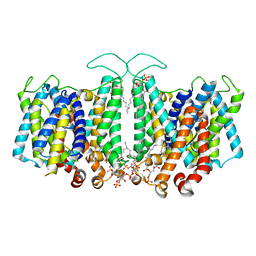 | | Cryo-EM Analysis of AE1 Structure in 100 mM NaHCO3 Buffer: Form2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Band 3 anion transport protein, DIUNDECYL PHOSPHATIDYL CHOLINE, ... | | Authors: | Su, C.C. | | Deposit date: | 2023-06-08 | | Release date: | 2024-05-22 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Cryo-EM structure of the band 3 anion transport protein
To Be Published
|
|
8T47
 
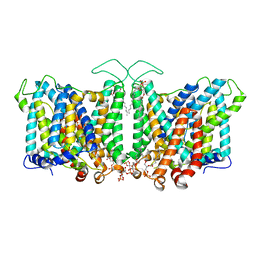 | | Cryo-EM Analysis of AE1 Structure in 100 mM NaHCO3 Buffer: Form3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Band 3 anion transport protein, CARBONATE ION, ... | | Authors: | Su, C.C. | | Deposit date: | 2023-06-08 | | Release date: | 2024-05-22 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Cryo-EM structure of the band 3 anion transport protein
To Be Published
|
|
8T44
 
 | | Cryo-EM Analysis of AE1 Structure in 100 mM NaHCO3 Buffer: Form1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Band 3 anion transport protein, CARBONATE ION, ... | | Authors: | Su, C.C. | | Deposit date: | 2023-06-08 | | Release date: | 2024-05-22 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Cryo-EM structure of the band 3 anion transport protein
To Be Published
|
|
8T3R
 
 | | Cryo-EM Analysis of AE1 Structure in 100 mM NaCl Buffer: Form1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Band 3 anion transport protein, CHLORIDE ION, ... | | Authors: | Su, C.C. | | Deposit date: | 2023-06-07 | | Release date: | 2024-05-22 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Cryo-EM structure of the band 3 anion transport protein
To Be Published
|
|
8EMR
 
 | | Cryo-EM structure of human liver glucosidase II | | Descriptor: | CALCIUM ION, Glucosidase 2 subunit beta, Neutral alpha-glucosidase AB, ... | | Authors: | Su, C, Lyu, M, Zhang, Z, Yu, E.W. | | Deposit date: | 2022-09-28 | | Release date: | 2023-05-10 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | High-resolution structural-omics of human liver enzymes.
Cell Rep, 42, 2023
|
|
8EMT
 
 | | Cryo-EM analysis of the human aldehyde oxidase from liver | | Descriptor: | Aldehyde oxidase, DIOXOTHIOMOLYBDENUM(VI) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Su, C, Lyu, M, Zhang, Z, Yu, E.W. | | Deposit date: | 2022-09-28 | | Release date: | 2023-05-10 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | High-resolution structural-omics of human liver enzymes.
Cell Rep, 42, 2023
|
|
8EMS
 
 | | Cryo-EM structure of human liver glycogen phosphorylase | | Descriptor: | Glycogen phosphorylase, liver form, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Su, C, Lyu, M, Zhang, Z, Yu, E.W. | | Deposit date: | 2022-09-28 | | Release date: | 2023-05-10 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | High-resolution structural-omics of human liver enzymes.
Cell Rep, 42, 2023
|
|
8EL9
 
 | | Cryo-EM structure of human catalase | | Descriptor: | Catalase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Su, C.C. | | Deposit date: | 2022-09-23 | | Release date: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (2.27 Å) | | Cite: | Cryo-EM structure of human catalase
To Be Published
|
|
3OOC
 
 | | Crystal structure of the membrane fusion protein CusB from Escherichia coli | | Descriptor: | Cation efflux system protein cusB | | Authors: | Su, C.-C, Yang, F, Long, F, Reyon, D, Routh, M.D, Kuo, D.W, Mokhtari, A.K, Van Ornam, J.D, Rabe, K.L, Hoy, J.A, Lee, Y.J, Rajashankar, K.R, Yu, E.W. | | Deposit date: | 2010-08-30 | | Release date: | 2010-12-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.404 Å) | | Cite: | Crystal structure of the membrane fusion protein CusB from Escherichia coli.
J.Mol.Biol., 393, 2009
|
|
4DNT
 
 | | Crystal structure of the CusBA heavy-metal efflux complex from Escherichia coli, mutant | | Descriptor: | Cation efflux system protein CusA, Cation efflux system protein CusB | | Authors: | Su, C.-C, Long, F, Yu, E. | | Deposit date: | 2012-02-09 | | Release date: | 2012-06-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Charged Amino Acids (R83, E567, D617, E625, R669, and K678) of CusA Are Required for Metal Ion Transport in the Cus Efflux System.
J.Mol.Biol., 422, 2012
|
|
4DOP
 
 | | Crystal structure of the CusBA heavy-metal efflux complex from Escherichia coli, R mutant | | Descriptor: | Cation efflux system protein CusA, Cation efflux system protein CusB | | Authors: | Su, C.-C, Long, F, Yu, E. | | Deposit date: | 2012-02-10 | | Release date: | 2012-06-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Charged Amino Acids (R83, E567, D617, E625, R669, and K678) of CusA Are Required for Metal Ion Transport in the Cus Efflux System.
J.Mol.Biol., 422, 2012
|
|
3OPO
 
 | |
5LQ3
 
 | |
3K07
 
 | | Crystal structure of CusA | | Descriptor: | Cation efflux system protein cusA | | Authors: | Su, C.-C. | | Deposit date: | 2009-09-24 | | Release date: | 2010-09-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.521 Å) | | Cite: | Crystal structures of the CusA efflux pump suggest methionine-mediated metal transport.
Nature, 467, 2010
|
|
6OR2
 
 | | MmpL3 is a lipid transporter that binds trehalose monomycolate and phosphatidylethanolamine | | Descriptor: | (1S)-2-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-1-[(octadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, DODECYL-BETA-D-MALTOSIDE, Membrane protein, ... | | Authors: | Su, C.-C. | | Deposit date: | 2019-04-29 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | MmpL3 is a lipid transporter that binds trehalose monomycolate and phosphatidylethanolamine.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
7K8D
 
 | |
7K8C
 
 | |
7K8A
 
 | |
