2OYM
 
 | | Endo-glycoceramidase II from Rhodococcus sp.: five-membered iminocyclitol complex | | 分子名称: | Endoglycoceramidase II, N-{[(2R,3R,4R,5R)-3,4-DIHYDROXY-5-(HYDROXYMETHYL)PYRROLIDIN-2-YL]METHYL}-4-(DIMETHYLAMINO)BENZAMIDE, SODIUM ION | | 著者 | Caines, M.E.C, Strynadka, N.C.J. | | 登録日 | 2007-02-22 | | 公開日 | 2007-03-06 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.86 Å) | | 主引用文献 | The structural basis of glycosidase inhibition by five-membered iminocyclitols: the clan a glycoside hydrolase endoglycoceramidase as a model system.
Angew.Chem.Int.Ed.Engl., 46, 2007
|
|
1SN8
 
 | | Crystal structure of the S1 domain of RNase E from E. coli (Pb derivative) | | 分子名称: | LEAD (II) ION, Ribonuclease E | | 著者 | Schubert, M, Edge, R.E, Lario, P, Cook, M.A, Strynadka, N.C.J, Mackie, G.A, McIntosh, L.P. | | 登録日 | 2004-03-10 | | 公開日 | 2004-08-17 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural characterization of the RNase E S1 domain and identification of its oligonucleotide-binding and dimerization interfaces.
J.Mol.Biol., 341, 2004
|
|
1SMX
 
 | | Crystal structure of the S1 domain of RNase E from E. coli (native) | | 分子名称: | Ribonuclease E | | 著者 | Schubert, M, Edge, R.E, Lario, P, Cook, M.A, Strynadka, N.C.J, Mackie, G.A, McIntosh, L.P. | | 登録日 | 2004-03-09 | | 公開日 | 2004-08-17 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural characterization of the RNase E S1 domain and identification of its oligonucleotide-binding and dimerization interfaces.
J.Mol.Biol., 341, 2004
|
|
4QDD
 
 | | Crystal structure of 3-ketosteroid-9-alpha-hydroxylase 5 (KshA5) from R. rhodochrous in complex with 1,4-30Q-CoA | | 分子名称: | 3-ketosteroid 9alpha-hydroxylase oxygenase, FE (II) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | 著者 | Penfield, J, Worrall, L.J, Strynadka, N.C, Eltis, L.D. | | 登録日 | 2014-05-13 | | 公開日 | 2014-07-30 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Substrate specificities and conformational flexibility of 3-ketosteroid 9 alpha-hydroxylases.
J.Biol.Chem., 289, 2014
|
|
1RO8
 
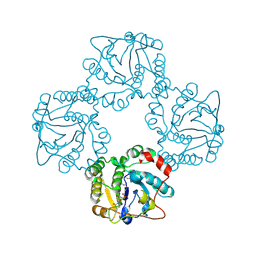 | | Structural analysis of the sialyltransferase CstII from Campylobacter jejuni in complex with a substrate analogue, cytidine-5'-monophosphate | | 分子名称: | CYTIDINE-5'-MONOPHOSPHATE, alpha-2,3/8-sialyltransferase | | 著者 | Chiu, C.P, Watts, A.G, Lairson, L.L, Gilbert, M, Lim, D, Wakarchuk, W.W, Withers, S.G, Strynadka, N.C. | | 登録日 | 2003-12-01 | | 公開日 | 2004-02-03 | | 最終更新日 | 2021-10-27 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Structural analysis of the sialyltransferase CstII from Campylobacter jejuni in complex with a substrate analog.
Nat.Struct.Mol.Biol., 11, 2004
|
|
4QDC
 
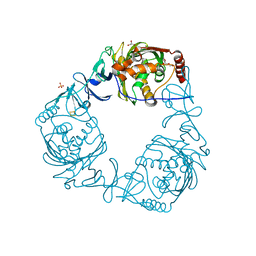 | | Crystal structure of 3-ketosteroid-9-alpha-hydroxylase 5 (KshA5) from R. rhodochrous in complex with FE2/S2 (INORGANIC) CLUSTER | | 分子名称: | 3-ketosteroid 9alpha-hydroxylase oxygenase, 4-ANDROSTENE-3-17-DIONE, FE (III) ION, ... | | 著者 | Penfield, J, Worrall, L.J, Strynadka, N.C, Eltis, L.D. | | 登録日 | 2014-05-13 | | 公開日 | 2014-07-30 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Substrate specificities and conformational flexibility of 3-ketosteroid 9 alpha-hydroxylases.
J.Biol.Chem., 289, 2014
|
|
4QCK
 
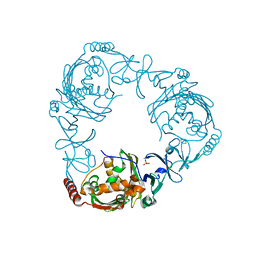 | | Crystal structure of 3-ketosteroid-9-alpha-hydroxylase (KshA) from M. tuberculosis in complex with 4-androstene-3,17-dione | | 分子名称: | 3-ketosteroid-9-alpha-monooxygenase oxygenase subunit, 4-ANDROSTENE-3-17-DIONE, FE (III) ION, ... | | 著者 | Penfield, J, Worrall, L.J, Strynadka, N.C, Eltis, L.D. | | 登録日 | 2014-05-12 | | 公開日 | 2014-07-30 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.46 Å) | | 主引用文献 | Substrate specificities and conformational flexibility of 3-ketosteroid 9 alpha-hydroxylases.
J.Biol.Chem., 289, 2014
|
|
4QDF
 
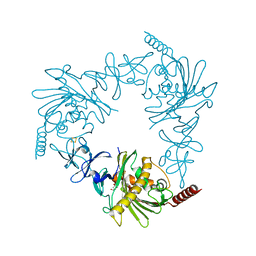 | | Crystal structure of apo KshA5 and KshA1 in complex with 1,4-30Q-CoA from R. rhodochrous | | 分子名称: | 3-ketosteroid 9alpha-hydroxylase oxygenase, FE (II) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | 著者 | Penfield, J, Worrall, L.J, Strynadka, N.C, Eltis, L.D. | | 登録日 | 2014-05-13 | | 公開日 | 2014-07-30 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.43 Å) | | 主引用文献 | Substrate specificities and conformational flexibility of 3-ketosteroid 9 alpha-hydroxylases.
J.Biol.Chem., 289, 2014
|
|
1RO7
 
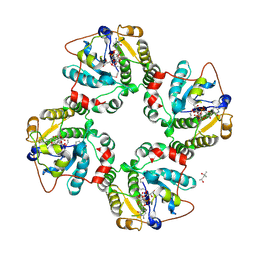 | | Structural analysis of the sialyltransferase CstII from Campylobacter jejuni in complex with a substrate analogue, CMP-3FNeuAc. | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, CYTIDINE-5'-MONOPHOSPHATE-3-FLUORO-N-ACETYL-NEURAMINIC ACID, alpha-2,3/8-sialyltransferase | | 著者 | Chiu, C.P, Watts, A.G, Lairson, L.L, Gilbert, M, Lim, D, Wakarchuk, W.W, Withers, S.G, Strynadka, N.C. | | 登録日 | 2003-12-01 | | 公開日 | 2004-02-03 | | 最終更新日 | 2021-10-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural analysis of the sialyltransferase CstII from Campylobacter jejuni in complex with a substrate analog.
Nat.Struct.Mol.Biol., 11, 2004
|
|
1SLJ
 
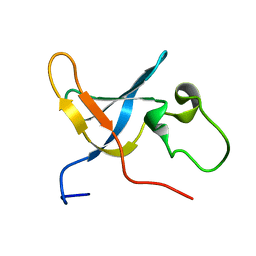 | | Solution structure of the S1 domain of RNase E from E. coli | | 分子名称: | Ribonuclease E | | 著者 | Schubert, M, Edge, R.E, Lario, P, Cook, M.A, Strynadka, N.C.J, Mackie, G.A, McIntosh, L.P. | | 登録日 | 2004-03-05 | | 公開日 | 2004-08-17 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural characterization of the RNase E S1 domain and identification of its oligonucleotide-binding and dimerization interfaces.
J.Mol.Biol., 341, 2004
|
|
3GR1
 
 | |
3GMY
 
 | |
7TC5
 
 | | All Phe-Azurin variant - F15Y | | 分子名称: | Azurin, COPPER (II) ION, NITRATE ION, ... | | 著者 | Fedoretz-Maxwell, B.P, Worrall, L.J, Strynadka, N.C.J, Warren, J.J. | | 登録日 | 2021-12-22 | | 公開日 | 2022-06-22 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | The Impact of Second Coordination Sphere Methionine-Aromatic Interactions in Copper Proteins.
Inorg.Chem., 61, 2022
|
|
7TC6
 
 | | All Phe-Azurin variant - F15W | | 分子名称: | Azurin, COPPER (II) ION, NITRATE ION | | 著者 | Fedoretz-Maxwell, B.P, Worrall, L.J, Strynadka, N.C.J, Warren, J.J. | | 登録日 | 2021-12-22 | | 公開日 | 2022-06-22 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | The Impact of Second Coordination Sphere Methionine-Aromatic Interactions in Copper Proteins.
Inorg.Chem., 61, 2022
|
|
3GMX
 
 | |
3GR0
 
 | |
3GMV
 
 | | Crystal Structure of Beta-Lactamse Inhibitory Protein-I (BLIP-I) in Apo Form | | 分子名称: | Beta-lactamase inhibitory protein BLIP-I, TRIS(HYDROXYETHYL)AMINOMETHANE | | 著者 | Lim, D.C, Gretes, M, Strynadka, N.C.J. | | 登録日 | 2009-03-15 | | 公開日 | 2009-03-31 | | 最終更新日 | 2017-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Insights into positive and negative requirements for protein-protein interactions by crystallographic analysis of the beta-lactamase inhibitory proteins BLIP, BLIP-I, and BLP.
J.Mol.Biol., 389, 2009
|
|
3GMW
 
 | | Crystal Structure of Beta-Lactamse Inhibitory Protein-I (BLIP-I) in Complex with TEM-1 Beta-Lactamase | | 分子名称: | B-lactamase, Beta-lactamase inhibitory protein BLIP-I, PHOSPHATE ION | | 著者 | Lim, D.C, Gretes, M, Strynadka, N.C.J. | | 登録日 | 2009-03-15 | | 公開日 | 2009-03-31 | | 最終更新日 | 2017-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Insights into positive and negative requirements for protein-protein interactions by crystallographic analysis of the beta-lactamase inhibitory proteins BLIP, BLIP-I, and BLP.
J.Mol.Biol., 389, 2009
|
|
3GR5
 
 | |
3HOE
 
 | |
3HOL
 
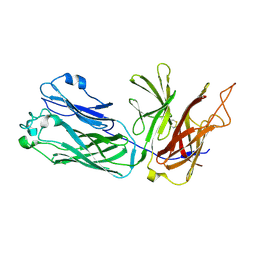 | |
3IR6
 
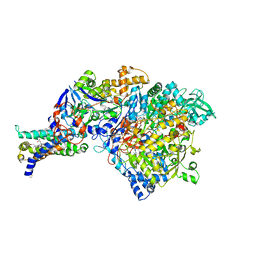 | | Crystal structure of NarGHI mutant NarG-H49S | | 分子名称: | (1S)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PENTANOYLOXY)METHYL]ETHYL OCTANOATE, FE3-S4 CLUSTER, GUANOSINE-5'-DIPHOSPHATE, ... | | 著者 | Bertero, M.G, Rothery, R.A, Weiner, J.H, Strynadka, N.C.J. | | 登録日 | 2009-08-21 | | 公開日 | 2010-01-05 | | 最終更新日 | 2021-10-13 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Protein crystallography reveals a role for the FS0 cluster of Escherichia coli nitrate reductase A (NarGHI) in enzyme maturation.
J.Biol.Chem., 285, 2010
|
|
3IR5
 
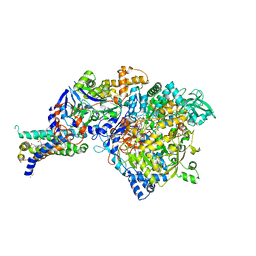 | | Crystal structure of NarGHI mutant NarG-H49C | | 分子名称: | (1S)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PENTANOYLOXY)METHYL]ETHYL OCTANOATE, FE3-S4 CLUSTER, IRON/SULFUR CLUSTER, ... | | 著者 | Bertero, M.G, Rothery, R.A, Weiner, J.H, Strynadka, N.C.J. | | 登録日 | 2009-08-21 | | 公開日 | 2010-01-05 | | 最終更新日 | 2021-10-13 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Protein crystallography reveals a role for the FS0 cluster of Escherichia coli nitrate reductase A (NarGHI) in enzyme maturation.
J.Biol.Chem., 285, 2010
|
|
3IR7
 
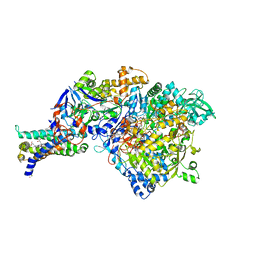 | | Crystal structure of NarGHI mutant NarG-R94S | | 分子名称: | (1S)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PENTANOYLOXY)METHYL]ETHYL OCTANOATE, FE3-S4 CLUSTER, IRON/SULFUR CLUSTER, ... | | 著者 | Bertero, M.G, Rothery, R.A, Weiner, J.H, Strynadka, N.C.J. | | 登録日 | 2009-08-21 | | 公開日 | 2010-01-05 | | 最終更新日 | 2021-10-13 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Protein crystallography reveals a role for the FS0 cluster of Escherichia coli nitrate reductase A (NarGHI) in enzyme maturation.
J.Biol.Chem., 285, 2010
|
|
3J1X
 
 | | A refined model of the prototypical Salmonella typhimurium T3SS basal body reveals the molecular basis for its assembly | | 分子名称: | Protein PrgH | | 著者 | Sgourakis, N.G, Bergeron, J.R.C, Worrall, L.J, Strynadka, N.C.J, Baker, D. | | 登録日 | 2012-07-10 | | 公開日 | 2013-05-22 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (11.7 Å) | | 主引用文献 | A Refined Model of the Prototypical Salmonella SPI-1 T3SS Basal Body Reveals the Molecular Basis for Its Assembly.
Plos Pathog., 9, 2013
|
|
