1SMX
 
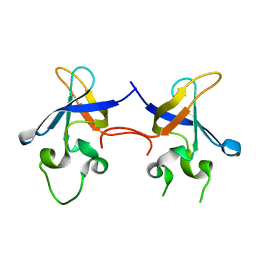 | | Crystal structure of the S1 domain of RNase E from E. coli (native) | | Descriptor: | Ribonuclease E | | Authors: | Schubert, M, Edge, R.E, Lario, P, Cook, M.A, Strynadka, N.C.J, Mackie, G.A, McIntosh, L.P. | | Deposit date: | 2004-03-09 | | Release date: | 2004-08-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural characterization of the RNase E S1 domain and identification of its oligonucleotide-binding and dimerization interfaces.
J.Mol.Biol., 341, 2004
|
|
1FOF
 
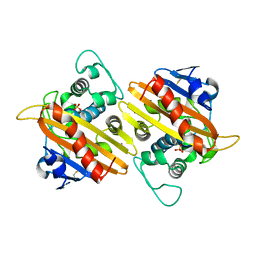 | | CRYSTAL STRUCTURE OF THE CLASS D BETA-LACTAMASE OXA-10 | | Descriptor: | BETA LACTAMASE OXA-10, COBALT (II) ION, SULFATE ION | | Authors: | Paetzel, M, Danel, F, de Castro, L, Mosimann, S.C, Page, M.G.P, Strynadka, N.C.J. | | Deposit date: | 2000-08-28 | | Release date: | 2000-10-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the class D beta-lactamase OXA-10.
Nat.Struct.Biol., 7, 2000
|
|
1RO8
 
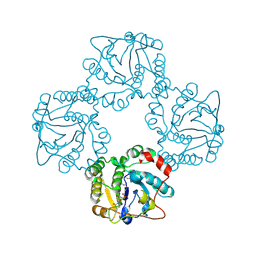 | | Structural analysis of the sialyltransferase CstII from Campylobacter jejuni in complex with a substrate analogue, cytidine-5'-monophosphate | | Descriptor: | CYTIDINE-5'-MONOPHOSPHATE, alpha-2,3/8-sialyltransferase | | Authors: | Chiu, C.P, Watts, A.G, Lairson, L.L, Gilbert, M, Lim, D, Wakarchuk, W.W, Withers, S.G, Strynadka, N.C. | | Deposit date: | 2003-12-01 | | Release date: | 2004-02-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural analysis of the sialyltransferase CstII from Campylobacter jejuni in complex with a substrate analog.
Nat.Struct.Mol.Biol., 11, 2004
|
|
1RO7
 
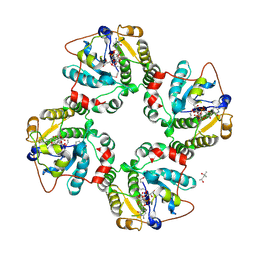 | | Structural analysis of the sialyltransferase CstII from Campylobacter jejuni in complex with a substrate analogue, CMP-3FNeuAc. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CYTIDINE-5'-MONOPHOSPHATE-3-FLUORO-N-ACETYL-NEURAMINIC ACID, alpha-2,3/8-sialyltransferase | | Authors: | Chiu, C.P, Watts, A.G, Lairson, L.L, Gilbert, M, Lim, D, Wakarchuk, W.W, Withers, S.G, Strynadka, N.C. | | Deposit date: | 2003-12-01 | | Release date: | 2004-02-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of the sialyltransferase CstII from Campylobacter jejuni in complex with a substrate analog.
Nat.Struct.Mol.Biol., 11, 2004
|
|
4QDC
 
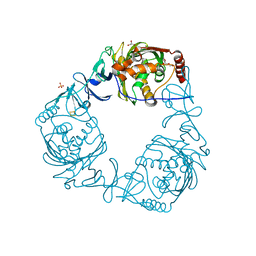 | | Crystal structure of 3-ketosteroid-9-alpha-hydroxylase 5 (KshA5) from R. rhodochrous in complex with FE2/S2 (INORGANIC) CLUSTER | | Descriptor: | 3-ketosteroid 9alpha-hydroxylase oxygenase, 4-ANDROSTENE-3-17-DIONE, FE (III) ION, ... | | Authors: | Penfield, J, Worrall, L.J, Strynadka, N.C, Eltis, L.D. | | Deposit date: | 2014-05-13 | | Release date: | 2014-07-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Substrate specificities and conformational flexibility of 3-ketosteroid 9 alpha-hydroxylases.
J.Biol.Chem., 289, 2014
|
|
4QCK
 
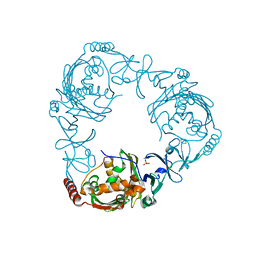 | | Crystal structure of 3-ketosteroid-9-alpha-hydroxylase (KshA) from M. tuberculosis in complex with 4-androstene-3,17-dione | | Descriptor: | 3-ketosteroid-9-alpha-monooxygenase oxygenase subunit, 4-ANDROSTENE-3-17-DIONE, FE (III) ION, ... | | Authors: | Penfield, J, Worrall, L.J, Strynadka, N.C, Eltis, L.D. | | Deposit date: | 2014-05-12 | | Release date: | 2014-07-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Substrate specificities and conformational flexibility of 3-ketosteroid 9 alpha-hydroxylases.
J.Biol.Chem., 289, 2014
|
|
4QDF
 
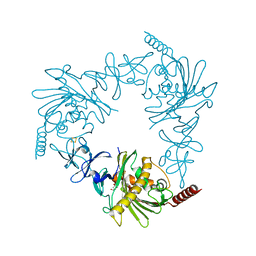 | | Crystal structure of apo KshA5 and KshA1 in complex with 1,4-30Q-CoA from R. rhodochrous | | Descriptor: | 3-ketosteroid 9alpha-hydroxylase oxygenase, FE (II) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Penfield, J, Worrall, L.J, Strynadka, N.C, Eltis, L.D. | | Deposit date: | 2014-05-13 | | Release date: | 2014-07-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Substrate specificities and conformational flexibility of 3-ketosteroid 9 alpha-hydroxylases.
J.Biol.Chem., 289, 2014
|
|
3GR1
 
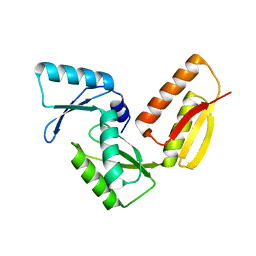 | |
3HOE
 
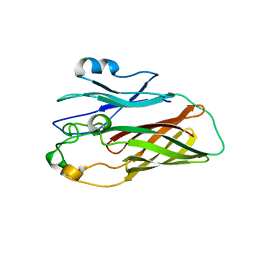 | |
3HOL
 
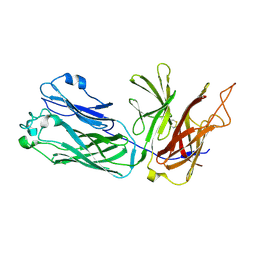 | |
3GMV
 
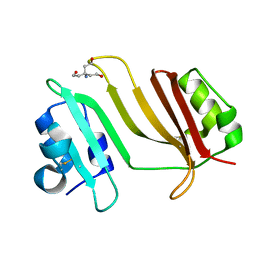 | | Crystal Structure of Beta-Lactamse Inhibitory Protein-I (BLIP-I) in Apo Form | | Descriptor: | Beta-lactamase inhibitory protein BLIP-I, TRIS(HYDROXYETHYL)AMINOMETHANE | | Authors: | Lim, D.C, Gretes, M, Strynadka, N.C.J. | | Deposit date: | 2009-03-15 | | Release date: | 2009-03-31 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Insights into positive and negative requirements for protein-protein interactions by crystallographic analysis of the beta-lactamase inhibitory proteins BLIP, BLIP-I, and BLP.
J.Mol.Biol., 389, 2009
|
|
1G68
 
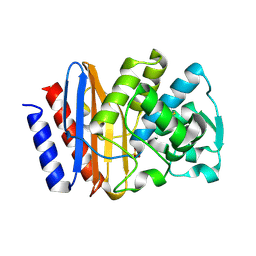 | | PSE-4 CARBENICILLINASE, WILD TYPE | | Descriptor: | BETA-LACTAMASE PSE-4, SULFATE ION | | Authors: | Lim, D, Sanschagrin, F, Passmore, L, De Castro, L, Levesque, R.C, Strynadka, N.C.J. | | Deposit date: | 2000-11-03 | | Release date: | 2001-02-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Insights into the molecular basis for the carbenicillinase activity of PSE-4 beta-lactamase from crystallographic and kinetic studies.
Biochemistry, 40, 2001
|
|
3GMX
 
 | |
1G6A
 
 | | PSE-4 CARBENICILLINASE, R234K MUTANT | | Descriptor: | BETA-LACTAMASE PSE-4, SULFATE ION | | Authors: | Lim, D, Sanschagrin, F, Passmore, L, De Castro, L, Levesque, R.C, Strynadka, N.C.J. | | Deposit date: | 2000-11-03 | | Release date: | 2001-02-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Insights into the molecular basis for the carbenicillinase activity of PSE-4 beta-lactamase from crystallographic and kinetic studies.
Biochemistry, 40, 2001
|
|
1G9R
 
 | | CRYSTAL STRUCTURE OF GALACTOSYLTRANSFERASE LGTC IN COMPLEX WITH MN AND UDP-2F-GALACTOSE | | Descriptor: | ACETIC ACID, GLYCOSYL TRANSFERASE, MANGANESE (II) ION, ... | | Authors: | Persson, K, Hoa, D.L, Diekelmann, M, Wakarchuk, W.W, Withers, S.G, Strynadka, N.C.J. | | Deposit date: | 2000-11-27 | | Release date: | 2001-02-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the retaining galactosyltransferase LgtC from Neisseria meningitidis in complex with donor and acceptor sugar analogs.
Nat.Struct.Biol., 8, 2001
|
|
1GA8
 
 | | CRYSTAL STRUCTURE OF GALACOSYLTRANSFERASE LGTC IN COMPLEX WITH DONOR AND ACCEPTOR SUGAR ANALOGS. | | Descriptor: | 4-deoxy-beta-D-xylo-hexopyranose-(1-4)-beta-D-glucopyranose, GALACTOSYL TRANSFERASE LGTC, MANGANESE (II) ION, ... | | Authors: | Persson, K, Ly, H.D, Diekelmann, M, Wakarchuk, W.W, Withers, S.G, Strynadka, N.C.J. | | Deposit date: | 2000-11-29 | | Release date: | 2001-02-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the retaining galactosyltransferase LgtC from Neisseria meningitidis in complex with donor and acceptor sugar analogs.
Nat.Struct.Biol., 8, 2001
|
|
3GMW
 
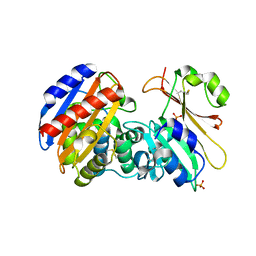 | | Crystal Structure of Beta-Lactamse Inhibitory Protein-I (BLIP-I) in Complex with TEM-1 Beta-Lactamase | | Descriptor: | B-lactamase, Beta-lactamase inhibitory protein BLIP-I, PHOSPHATE ION | | Authors: | Lim, D.C, Gretes, M, Strynadka, N.C.J. | | Deposit date: | 2009-03-15 | | Release date: | 2009-03-31 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Insights into positive and negative requirements for protein-protein interactions by crystallographic analysis of the beta-lactamase inhibitory proteins BLIP, BLIP-I, and BLP.
J.Mol.Biol., 389, 2009
|
|
3GR5
 
 | |
3IR6
 
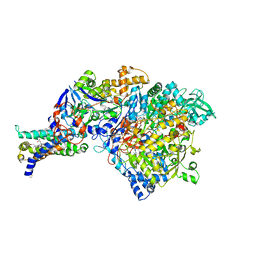 | | Crystal structure of NarGHI mutant NarG-H49S | | Descriptor: | (1S)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PENTANOYLOXY)METHYL]ETHYL OCTANOATE, FE3-S4 CLUSTER, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Bertero, M.G, Rothery, R.A, Weiner, J.H, Strynadka, N.C.J. | | Deposit date: | 2009-08-21 | | Release date: | 2010-01-05 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Protein crystallography reveals a role for the FS0 cluster of Escherichia coli nitrate reductase A (NarGHI) in enzyme maturation.
J.Biol.Chem., 285, 2010
|
|
1JHF
 
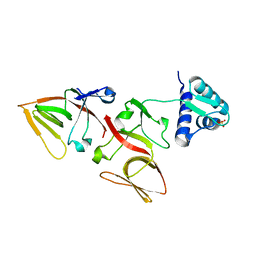 | | LEXA G85D MUTANT | | Descriptor: | LEXA REPRESSOR, SULFATE ION | | Authors: | Luo, Y, Pfuetzner, R.A, Mosimann, S, Little, J.W, Strynadka, N.C.J. | | Deposit date: | 2001-06-27 | | Release date: | 2001-09-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of LexA: a conformational switch for regulation of self-cleavage.
Cell(Cambridge,Mass.), 106, 2001
|
|
3GR0
 
 | |
3GMY
 
 | |
1JDI
 
 | | CRYSTAL STRUCTURE OF L-RIBULOSE-5-PHOSPHATE 4-EPIMERASE | | Descriptor: | L-RIBULOSE 5 PHOSPHATE 4-EPIMERASE, ZINC ION | | Authors: | Luo, Y, Samuel, J, Mosimann, S.C, Lee, J.E, Tanner, M.E, Strynadka, N.C.J. | | Deposit date: | 2001-06-13 | | Release date: | 2002-01-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structure of L-ribulose-5-phosphate 4-epimerase: an aldolase-like platform for epimerization.
Biochemistry, 40, 2001
|
|
3IR5
 
 | | Crystal structure of NarGHI mutant NarG-H49C | | Descriptor: | (1S)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PENTANOYLOXY)METHYL]ETHYL OCTANOATE, FE3-S4 CLUSTER, IRON/SULFUR CLUSTER, ... | | Authors: | Bertero, M.G, Rothery, R.A, Weiner, J.H, Strynadka, N.C.J. | | Deposit date: | 2009-08-21 | | Release date: | 2010-01-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Protein crystallography reveals a role for the FS0 cluster of Escherichia coli nitrate reductase A (NarGHI) in enzyme maturation.
J.Biol.Chem., 285, 2010
|
|
1JHH
 
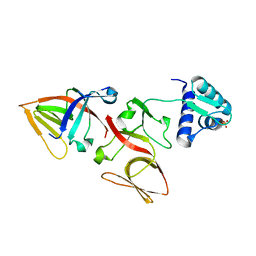 | | LEXA S119A MUTANT | | Descriptor: | LEXA REPRESSOR, SULFATE ION | | Authors: | Luo, Y, Pfuetzner, R.A, Mosimann, S, Little, J.W, Strynadka, N.C.J. | | Deposit date: | 2001-06-27 | | Release date: | 2001-09-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of LexA: a conformational switch for regulation of self-cleavage.
Cell(Cambridge,Mass.), 106, 2001
|
|
