1OQ6
 
 | | solution structure of Copper-S46V CopA from Bacillus subtilis | | Descriptor: | COPPER (II) ION, Potential copper-transporting ATPase | | Authors: | Banci, L, Bertini, I, Ciofi-Baffoni, S, Gonnelli, l, Su, X.C, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2003-03-07 | | Release date: | 2003-09-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A core mutation affecting the folding properties of a soluble domain of the ATPase protein CopA from Bacillus subtilis
J.Mol.Biol., 331, 2003
|
|
1P6Q
 
 | | NMR Structure of the Response regulator CheY2 from Sinorhizobium meliloti, complexed with Mg++ | | Descriptor: | CheY2 | | Authors: | Riepl, H, Scharf, B, Maurer, T, Schmitt, R, Kalbitzer, H.R, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2003-04-30 | | Release date: | 2004-06-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the inactive and BeF3-activated response regulator CheY2.
J.Mol.Biol., 338, 2004
|
|
1P6U
 
 | | NMR structure of the BeF3-activated structure of the response regulator Chey2-Mg2+ from Sinorhizobium meliloti | | Descriptor: | CheY2 | | Authors: | Riepl, H, Scharf, B, Maurer, T, Schmitt, R, Kalbitzer, H.R, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2003-04-30 | | Release date: | 2003-11-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structures of the Inactive and BeF(3)-activated Response Regulator CheY2
J.Biol.Chem., 338, 2004
|
|
1OQ3
 
 | | A core mutation affecting the folding properties of a soluble domain of the ATPase protein CopA from Bacillus subtilis | | Descriptor: | Potential copper-transporting ATPase | | Authors: | Banci, L, Bertini, I, Ciofi-Baffoni, S, Gonnelli, L, Su, X.C, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2003-03-07 | | Release date: | 2003-09-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A core mutation affecting the folding properties of a soluble domain of the ATPase protein CopA from Bacillus subtilis
J.Mol.Biol., 331, 2003
|
|
2BDH
 
 | | Human Kallikrein 4 complex with zinc and p-aminobenzamidine | | Descriptor: | Kallikrein-4, P-AMINO BENZAMIDINE, ZINC ION | | Authors: | Debela, M, Bode, W, Goettig, P, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2005-10-20 | | Release date: | 2006-10-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of human tissue kallikrein 4: activity modulation by a specific zinc binding site.
J.Mol.Biol., 362, 2006
|
|
1PFJ
 
 | | Solution structure of the N-terminal PH/PTB domain of the TFIIH P62 subunit | | Descriptor: | TFIIH basal transcription factor complex p62 subunit | | Authors: | Gervais, V, Lamour, V, Jawhari, A, Frindel, F, Wasielewski, E, Thierry, J.C, Kieffer, B, Poterszman, A, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2003-05-27 | | Release date: | 2004-06-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | TFIIH contains a PH domain involved in DNA nucleotide excision repair.
Nat.Struct.Mol.Biol., 11, 2004
|
|
2BDI
 
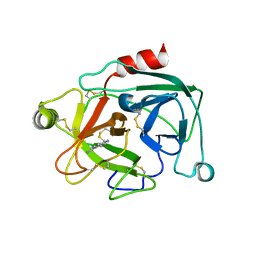 | | Human Kallikrein 4 complex with cobalt and p-aminobenzamidine | | Descriptor: | COBALT (II) ION, Kallikrein-4, P-AMINO BENZAMIDINE | | Authors: | Debela, M, Bode, W, Goettig, P, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2005-10-20 | | Release date: | 2006-10-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of human tissue kallikrein 4: activity modulation by a specific zinc binding site.
J.Mol.Biol., 362, 2006
|
|
2GA7
 
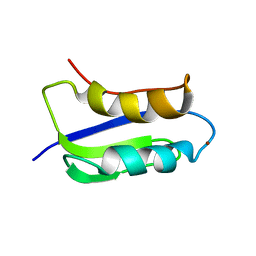 | | Solution structure of the copper(I) form of the third metal-binding domain of ATP7A protein (menkes disease protein) | | Descriptor: | COPPER (I) ION, Copper-transporting ATPase 1 | | Authors: | Banci, L, Bertini, I, Cantini, F, DellaMalva, N, Rosato, A, Herrmann, T, Wuthrich, K, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2006-03-08 | | Release date: | 2006-08-01 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure and intermolecular interactions of the third metal-binding domain of ATP7A, the Menkes disease protein.
J.Biol.Chem., 281, 2006
|
|
2FWV
 
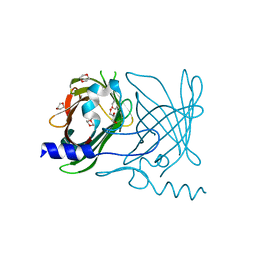 | | Crystal Structure of Rv0813 | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, GLYCEROL, hypothetical protein MtubF_01000852 | | Authors: | Shepard, W, Haouz, A, Grana, M, Buschiazzo, A, Betton, J.M, Cole, S.T, Alzari, P.M, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2006-02-03 | | Release date: | 2006-08-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Crystal Structure of Rv0813c from Mycobacterium tuberculosis Reveals a New Family of Fatty Acid-Binding Protein-Like Proteins in Bacteria
J.Bacteriol., 189, 2007
|
|
2BDG
 
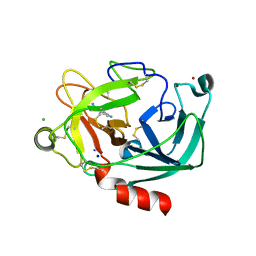 | | Human Kallikrein 4 complex with nickel and p-aminobenzamidine | | Descriptor: | CHLORIDE ION, Kallikrein-4, NICKEL (II) ION, ... | | Authors: | Debela, M, Bode, W, Goettig, P, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2005-10-20 | | Release date: | 2006-10-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structures of human tissue kallikrein 4: activity modulation by a specific zinc binding site.
J.Mol.Biol., 362, 2006
|
|
1Q0R
 
 | | Crystal structure of aclacinomycin methylesterase (RdmC) with bound product analogue, 10-decarboxymethylaclacinomycin T (DcmaT) | | Descriptor: | 10-DECARBOXYMETHYLACLACINOMYCIN T (DCMAT), PENTAETHYLENE GLYCOL, SULFATE ION, ... | | Authors: | Jansson, A, Niemi, J, Mantsala, P, Schneider, G, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2003-07-17 | | Release date: | 2003-11-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of aclacinomycin methylesterase with bound product analogues: implications for anthracycline recognition and mechanism.
J.Biol.Chem., 278, 2003
|
|
2GHF
 
 | |
1Q0Z
 
 | | Crystal structure of aclacinomycin methylesterase (RdmC) with bound product analogue, 10-decarboxymethylaclacinomycin A (DcmA) | | Descriptor: | 10-DECARBOXYMETHYLACLACINOMYCIN A (DCMAA), PENTAETHYLENE GLYCOL, SULFATE ION, ... | | Authors: | Jansson, A, Niemi, J, Mantsala, P, Schneider, G, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2003-07-18 | | Release date: | 2003-11-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of aclacinomycin methylesterase with bound product analogues: implications for anthracycline recognition and mechanism.
J.Biol.Chem., 278, 2003
|
|
2GQK
 
 | | Solution structure of Human Ni(II)-Sco1 | | Descriptor: | NICKEL (II) ION, SCO1 protein homolog, mitochondrial | | Authors: | Banci, L, Bertini, I, Calderone, V, Ciofi-Baffoni, S, Mangani, S, Palumaa, P, Martinelli, M, Wang, S, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2006-04-21 | | Release date: | 2006-06-06 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | A hint for the function of human Sco1 from different structures.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2GVP
 
 | | Solution structure of Human apo Sco1 | | Descriptor: | SCO1 protein homolog, mitochondrial | | Authors: | Banci, L, Bertini, I, Calderone, V, Ciofi-Baffoni, S, Mangani, S, Paulmaa, P, Martinelli, M, Wang, S, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2006-05-03 | | Release date: | 2006-06-06 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | A hint for the function of human Sco1 from different structures.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2HRN
 
 | | Solution Structure of Cu(I) P174L-HSco1 | | Descriptor: | COPPER (I) ION, SCO1 protein homolog, mitochondrial | | Authors: | Banci, L, Bertini, I, Ciofi-Baffoni, S, Leontari, I, Martinelli, M, Palumaa, P, Sillard, R, Wang, S, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2006-07-20 | | Release date: | 2007-01-16 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Human Sco1 functional studies and pathological implications of the P174L mutant.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2HRF
 
 | | Solution Structure of Cu(I) P174L HSco1 | | Descriptor: | COPPER (I) ION, SCO1 protein homolog, mitochondrial | | Authors: | Banci, L, Bertini, I, Ciofi-Baffoni, S, Leontari, I, Martinelli, M, Palumaa, P, Sillard, R, Wang, S, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2006-07-20 | | Release date: | 2007-01-16 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Human Sco1 functional studies and pathological implications of the P174L mutant.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
1NAQ
 
 | | Crystal structure of CUTA1 from E.coli at 1.7 A resolution | | Descriptor: | MERCURIBENZOIC ACID, MERCURY (II) ION, Periplasmic divalent cation tolerance protein cutA | | Authors: | Calderone, V, Mangani, S, Benvenuti, M, Viezzoli, M.S, Banci, L, Bertini, I, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2002-11-28 | | Release date: | 2003-11-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The evolutionarily conserved trimeric structure of CutA1 proteins suggests a role in signal transduction.
J.Biol.Chem., 278, 2003
|
|
2K1R
 
 | | The solution NMR structure of the complex between MNK1 and HAH1 mediated by Cu(I) | | Descriptor: | COPPER (II) ION, Copper transport protein ATOX1, Copper-transporting ATPase 1 | | Authors: | Bertini, I, Banci, L.C, Felli, I.C, Pavelkova, A, Rosato, A, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2008-03-14 | | Release date: | 2009-03-31 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the copper(I)-mediated complex between the first soluble domain of the Menkes protein and the metallochaperone HAH1.
To be Published
|
|
2GQM
 
 | | Solution structure of Human Cu(I)-Sco1 | | Descriptor: | COPPER (I) ION, SCO1 protein homolog, mitochondrial | | Authors: | Banci, L, Bertini, I, Calderone, V, Ciofi-Baffoni, S, Mangani, S, Palumaa, P, Martinelli, M, Wang, S, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2006-04-21 | | Release date: | 2006-06-06 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | A hint for the function of human Sco1 from different structures.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2JJX
 
 | | THE CRYSTAL STRUCTURE OF UMP KINASE FROM BACILLUS ANTHRACIS (BA1797) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, URIDYLATE KINASE | | Authors: | Meier, C, Carter, L.G, Mancini, E.J, Owens, R.J, Stuart, D.I, Esnouf, R.M, Oxford Protein Production Facility (OPPF), Structural Proteomics in Europe (SPINE) | | Deposit date: | 2008-04-23 | | Release date: | 2008-07-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | The Crystal Structure of Ump Kinase from Bacillus Anthracis (Ba1797) Reveals an Allosteric Nucleotide-Binding Site.
J.Mol.Biol., 381, 2008
|
|
1MB3
 
 | | CRYSTAL STRUCTURE OF THE RESPONSE REGULATOR DIVK AT PH 8.5 IN COMPLEX WITH MG2+ | | Descriptor: | MAGNESIUM ION, cell division response regulator DivK | | Authors: | Guillet, V, Ohta, N, Cabantous, S, Newton, A, Samama, J.-P, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2002-08-02 | | Release date: | 2002-12-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Crystallographic and Biochemical Studies of DivK Reveal Novel Features of
an Essential Response Regulator in Caulobacter crescentus.
J.Biol.Chem., 277, 2002
|
|
2G6V
 
 | | The crystal structure of ribD from Escherichia coli | | Descriptor: | Riboflavin biosynthesis protein ribD | | Authors: | Stenmark, P, Moche, M, Gurmu, D, Nordlund, P, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2006-02-25 | | Release date: | 2007-02-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Crystal Structure of the Bifunctional Deaminase/Reductase RibD of the Riboflavin Biosynthetic Pathway in Escherichia coli: Implications for the Reductive Mechanism.
J.Mol.Biol., 373, 2007
|
|
2GGP
 
 | | Solution structure of the Atx1-Cu(I)-Ccc2a complex | | Descriptor: | COPPER (I) ION, Metal homeostasis factor ATX1, Probable copper-transporting ATPase | | Authors: | Banci, L, Bertini, I, Cantini, F, Felli, I.C, Gonnelli, L, Hadjiliadis, N, Pierattelli, R, Rosato, A, Voulgaris, P, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2006-03-24 | | Release date: | 2006-08-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The Atx1-Ccc2 complex is a metal-mediated protein-protein interaction.
Nat.Chem.Biol., 2, 2006
|
|
1NM4
 
 | | Solution structure of Cu(I)-CopC from Pseudomonas syringae | | Descriptor: | Copper resistance protein C | | Authors: | Arnesano, F, Banci, L, Bertini, I, Mangani, S, Thompsett, A.R, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2003-01-09 | | Release date: | 2003-04-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A redox switch in CopC: An intriguing copper trafficking protein that binds copper(I) and copper(II)
at different sites
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
