5JRA
 
 | | Nitric oxide complex of the L16V mutant of cytochrome c prime from Alcaligenes xylosoxidans | | Descriptor: | ASCORBIC ACID, Cytochrome c', HEME C, ... | | Authors: | Kekilli, D, Strange, R.W, Hough, M.A. | | Deposit date: | 2016-05-06 | | Release date: | 2017-03-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Engineering proximal vs. distal heme-NO coordination via dinitrosyl dynamics: implications for NO sensor design.
Chem Sci, 8, 2017
|
|
7ADF
 
 | | SFX structure of dehaloperoxidase B in the ferric form | | Descriptor: | Dehaloperoxidase B, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Moreno Chicano, T, Ebrahim, A.E, Axford, D.A, Sherrell, D.A, Sugimoto, H, Tono, K, Owada, S, Worrall, J.W, Strange, R.W, Owen, R.L, Hough, M.A. | | Deposit date: | 2020-09-14 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | SFX structure of dehaloperoxidase B in the ferric form
to be published
|
|
7ACP
 
 | | Serial synchrotron structure of dehaloperoxidase B | | Descriptor: | Dehaloperoxidase B, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Moreno Chicano, T, Ebrahim, A.E, Axford, D.A, Sherrell, D.A, Worrall, J.W, Strange, R.W, Owen, R.L, Hough, M.A. | | Deposit date: | 2020-09-11 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | SFX structure of dehaloperoxidase B in the ferric form
to be published
|
|
5JS5
 
 | | Nitric oxide complex of the L16F mutant of cytochrome c prime from Alcaligenes xylosoxidans | | Descriptor: | ASCORBIC ACID, Cytochrome c', HEME C, ... | | Authors: | Kekilli, D, Strange, R.W, Hough, M.A. | | Deposit date: | 2016-05-07 | | Release date: | 2017-03-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Engineering proximal vs. distal heme-NO coordination via dinitrosyl dynamics: implications for NO sensor design.
Chem Sci, 8, 2017
|
|
1OBF
 
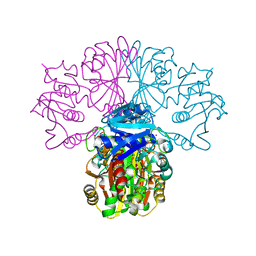 | | The crystal structure of Glyceraldehyde 3-phosphate Dehydrogenase from Alcaligenes xylosoxidans at 1.7A resolution. | | Descriptor: | GLYCERALDEHYDE 3-PHOSPHATE DEHYDROGENASE, POTASSIUM ION, SULFATE ION, ... | | Authors: | Antonyuk, S.V, Eady, R.R, Strange, R.W, Hasnain, S.S. | | Deposit date: | 2003-01-30 | | Release date: | 2003-06-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Structure of Glyceraldehyde 3-Phosphate Dehydrogenase from Alcaligenes Xylosoxidans at 1.7 A Resolution
Acta Crystallogr.,Sect.D, 59, 2003
|
|
5JLI
 
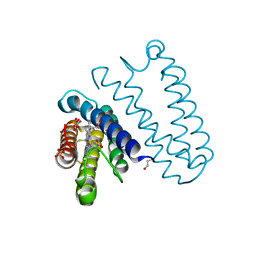 | | Nitric oxide complex of the L16A mutant of cytochrome c prime from Alcaligenes xylosoxidans | | Descriptor: | ASCORBIC ACID, Cytochrome c', HEME C, ... | | Authors: | Kekilli, D, Strange, R.W, Hough, M.A. | | Deposit date: | 2016-04-27 | | Release date: | 2017-03-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Engineering proximal vs. distal heme-NO coordination via dinitrosyl dynamics: implications for NO sensor design.
Chem Sci, 8, 2017
|
|
5AGF
 
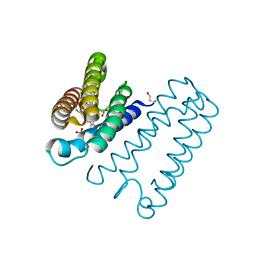 | | Nitrosyl complex of the D121Q variant of cytochrome c prime from Alcaligenes xylosoxidans | | Descriptor: | CYTOCHROME C PRIME, HEME C, NITRIC OXIDE, ... | | Authors: | Gahfoor, D.D, Kekilli, D, Abdullah, G.H, Dworkowski, F.S.N, Hassan, H.G, Wilson, M.T, Hough, M.A, Strange, R.W. | | Deposit date: | 2015-01-30 | | Release date: | 2015-09-09 | | Last modified: | 2020-03-11 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Hydrogen Bonding of the Dissociated Histidine Ligand is not Required for Formation of a Proximal No Adduct in Cytochrome C'.
J.Biol.Inorg.Chem., 20, 2015
|
|
1GS8
 
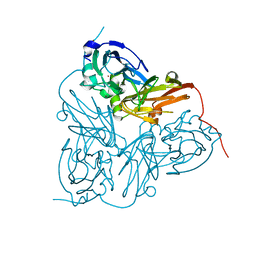 | | Crystal structure of mutant D92N Alcaligenes xylosoxidans Nitrite Reductase | | Descriptor: | COPPER (II) ION, DISSIMILATORY COPPER-CONTAINING NITRITE REDUCTASE, ZINC ION | | Authors: | Ellis, M.J, Prudencio, M, Dodd, F.E, Strange, R.W, Sawers, G, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2002-01-02 | | Release date: | 2002-02-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Biochemical and Crystallographic Studies of the met144Ala, Asp92Asn and His254Phe Mutants of the Nitrite Reductase from Alcaligenes Xylosoxidans Provide Insight Into the Enzyme Mechanism.
J.Mol.Biol., 316, 2002
|
|
3ZK4
 
 | |
6EWM
 
 | | Crystal structure of heme free PORPHYROMONAS GINGIVALIS HEME-BINDING PROTEIN HMUY | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, Haemophore HmuY, ... | | Authors: | Antonyuk, S.V, Strange, R.W, Bielecki, M, Olczak, T, Olczak, M. | | Deposit date: | 2017-11-05 | | Release date: | 2018-10-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Tannerella forsythiaTfo belongs toPorphyromonas gingivalisHmuY-like family of proteins but differs in heme-binding properties.
Biosci. Rep., 38, 2018
|
|
6EU8
 
 | |
5JVE
 
 | |
5JSL
 
 | | The L16F mutant of cytochrome c prime from Alcaligenes xylosoxidans: Ferrous form | | Descriptor: | ASCORBIC ACID, Cytochrome c', HEME C, ... | | Authors: | Kekilli, D, Strange, R.W, Hough, M.A. | | Deposit date: | 2016-05-08 | | Release date: | 2017-03-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Engineering proximal vs. distal heme-NO coordination via dinitrosyl dynamics: implications for NO sensor design.
Chem Sci, 8, 2017
|
|
5JT4
 
 | | L16A mutant of cytochrome c prime from Alcaligenes xylosoxidans: Ferrous state | | Descriptor: | ASCORBIC ACID, Cytochrome c', HEME C, ... | | Authors: | Kekilli, D, Strange, R.W, Hough, M.A. | | Deposit date: | 2016-05-09 | | Release date: | 2017-03-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Engineering proximal vs. distal heme-NO coordination via dinitrosyl dynamics: implications for NO sensor design.
Chem Sci, 8, 2017
|
|
5JUA
 
 | |
6GBY
 
 | | Copper nitrite reductase from Achromobacter cycloclastes: non-polymorph separated dataset 1 | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, NITRITE ION | | Authors: | Ebrahim, A, Appleby, M.V, Axford, D, Beale, J, Moreno-Chicano, T, Sherrell, D.A, Strange, R.W, Owen, R.L, Hough, M.A. | | Deposit date: | 2018-04-16 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Resolving polymorphs and radiation-driven effects in microcrystals using fixed-target serial synchrotron crystallography.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
5I6O
 
 | | Crystal Structure of Copper Nitrite Reductase at 100K after 20.70 MGy | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, NITRIC OXIDE, ... | | Authors: | Horrell, S, Hough, M.A, Strange, R.W. | | Deposit date: | 2016-02-16 | | Release date: | 2016-07-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Serial crystallography captures enzyme catalysis in copper nitrite reductase at atomic resolution from one crystal.
Iucrj, 3, 2016
|
|
5I6M
 
 | | Crystal Structure of Copper Nitrite Reductase at 100K after 7.59 MGy | | Descriptor: | ACETATE ION, COPPER (II) ION, Copper-containing nitrite reductase, ... | | Authors: | Horrell, S, Hough, M.A, Strange, R.W. | | Deposit date: | 2016-02-16 | | Release date: | 2016-07-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Serial crystallography captures enzyme catalysis in copper nitrite reductase at atomic resolution from one crystal.
Iucrj, 3, 2016
|
|
5I6P
 
 | |
5JP7
 
 | |
5I6L
 
 | | Crystal Structure of Copper Nitrite Reductase at 100K after 2.76 MGy | | Descriptor: | ACETATE ION, COPPER (II) ION, Copper-containing nitrite reductase, ... | | Authors: | Horrell, S, Hough, M.A, Strange, R.W. | | Deposit date: | 2016-02-16 | | Release date: | 2016-07-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Serial crystallography captures enzyme catalysis in copper nitrite reductase at atomic resolution from one crystal.
Iucrj, 3, 2016
|
|
5I6K
 
 | |
5I6N
 
 | | Crystal Structure of Copper Nitrite Reductase at 100K after 11.73 MGy | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, NITRIC OXIDE, ... | | Authors: | Horrell, S, Hough, M.A, Strange, R.W. | | Deposit date: | 2016-02-16 | | Release date: | 2016-07-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Serial crystallography captures enzyme catalysis in copper nitrite reductase at atomic resolution from one crystal.
Iucrj, 3, 2016
|
|
2WKO
 
 | | Structure of metal loaded Pathogenic SOD1 Mutant G93A. | | Descriptor: | COPPER (II) ION, IODIDE ION, SUPEROXIDE DISMUTASE [CU-ZN], ... | | Authors: | Antonyuk, S.V, Galaleldeen, A, Strange, R, Whitson, L, Narayana, N, Taylor, A, Schuermann, J.P, Holloway, S.P, Hasnain, S.S, Hart, P.J. | | Deposit date: | 2009-06-16 | | Release date: | 2009-11-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural and Biophysical Properties of Metal-Free Pathogenic Sod1 Mutants A4V and G93A.
Arch.Biochem.Biophys., 492, 2009
|
|
2BP0
 
 | | M144L mutant of nitrite reductase from Alcaligenes xylosoxidans | | Descriptor: | COPPER (II) ION, DISSIMILATORY COPPER-CONTAINING NITRITE REDUCTASE, SULFATE ION, ... | | Authors: | Hough, M.A, Ellis, M.J, Antonyuk, S, Strange, R.W, Sawers, G, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2005-04-17 | | Release date: | 2006-07-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High Resolution Structural Studies of Mutants Provide Insights Into Catalysis and Electron Transfer Processes in Copper Nitrite Reductase
J.Mol.Biol., 350, 2005
|
|
