1NEP
 
 | | Crystal Structure Analysis of the Bovine NPC2 (Niemann-Pick C2) Protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Epididymal secretory protein E1, PHOSPHATE ION | | Authors: | Friedland, N, Liou, H.-L, Lobel, P, Stock, A.M. | | Deposit date: | 2002-12-11 | | Release date: | 2003-01-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of a Cholesterol-binding Protein Deficient in Niemann-Pick Type C2 Disease
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
3EDY
 
 | | Crystal Structure of the Precursor Form of Human Tripeptidyl-Peptidase 1 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Guhaniyogi, J, Sohar, I, Das, K, Lobel, P, Stock, A.M. | | Deposit date: | 2008-09-03 | | Release date: | 2008-11-18 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure and Autoactivation Pathway of the Precursor Form of Human Tripeptidyl-peptidase 1, the Enzyme Deficient in Late Infantile Ceroid Lipofuscinosis
J.Biol.Chem., 284, 2009
|
|
4PA0
 
 | | Omecamtiv Mercarbil binding site on the Human Beta-Cardiac Myosin Motor Domain | | Descriptor: | GLYCEROL, Myosin-7,Green fluorescent protein, methyl 4-(2-fluoro-3-{[(6-methylpyridin-3-yl)carbamoyl]amino}benzyl)piperazine-1-carboxylate | | Authors: | Winkelmann, D.A, Miller, M.T, Stock, A.M. | | Deposit date: | 2014-04-06 | | Release date: | 2015-07-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis for drug-induced allosteric changes to human beta-cardiac myosin motor activity.
Nat Commun, 6, 2015
|
|
4P7H
 
 | |
1OPC
 
 | |
1DZ9
 
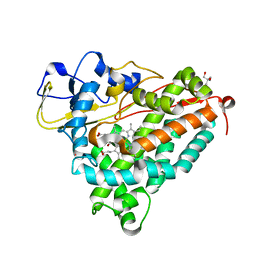 | | Putative oxo complex of P450cam from Pseudomonas putida | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CAMPHOR, CYTOCHROME P450-CAM, ... | | Authors: | Schlichting, I, Berendzen, J, Chu, K, Stock, A.M, Maves, S.A, Benson, D.E, Sweet, R.M, Ringe, D, Petsko, G.A, Sligar, S.G. | | Deposit date: | 2000-02-18 | | Release date: | 2000-03-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Catalytic Pathway of Cytochrome P450Cam at Atomic Resolution
Science, 287, 2000
|
|
4G4K
 
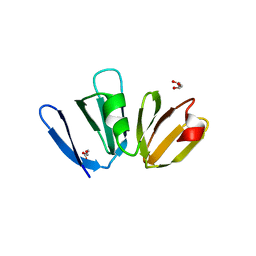 | |
1DZ4
 
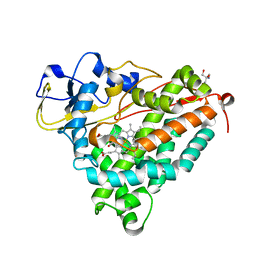 | | ferric p450cam from pseudomonas putida | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CAMPHOR, CYTOCHROME P450-CAM, ... | | Authors: | Schlichting, I, Berendzen, J, Chu, K, Stock, A.M, Maves, S.A, Benson, D.E, Sweet, R.M, Ringe, D, Petsko, G.A, Sligar, S.G. | | Deposit date: | 2000-02-16 | | Release date: | 2000-07-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Catalytic Pathway of Cytochrome P450Cam at Atomic Resolution
Science, 287, 2000
|
|
1DZ8
 
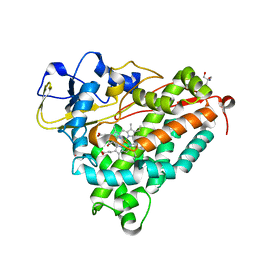 | | oxygen complex of p450cam from pseudomonas putida | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CAMPHOR, CYTOCHROME P450-CAM, ... | | Authors: | Schlichting, I, Berendzen, J, Chu, K, Stock, A.M, Maves, S.A, Benson, D.E, Sweet, R.M, Ringe, D, Petsko, G.A, Sligar, S.G. | | Deposit date: | 2000-02-18 | | Release date: | 2000-03-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Catalytic Pathway of Cytochrome P450Cam at Atomic Resolution
Science, 287, 2000
|
|
1DZ6
 
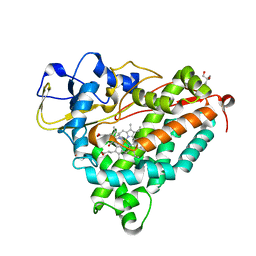 | | ferrous p450cam from pseudomonas putida | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CAMPHOR, CYTOCHROME P450-CAM, ... | | Authors: | Schlichting, I, Berendzen, J, Chu, K, Stock, A.M, Maves, S.A, Benson, D.E, Sweet, R.M, Ringe, D, Petsko, G.A, Sligar, S.G. | | Deposit date: | 2000-02-18 | | Release date: | 2000-03-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Catalytic Pathway of Cytochrome P450Cam at Atomic Resolution
Science, 287, 2000
|
|
4IF4
 
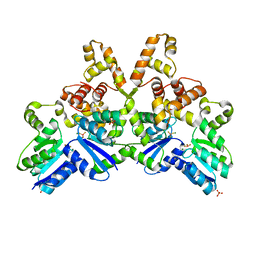 | |
4GVP
 
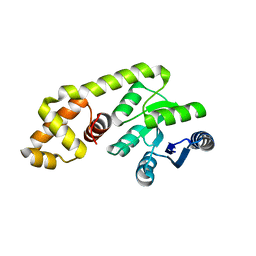 | |
3NNN
 
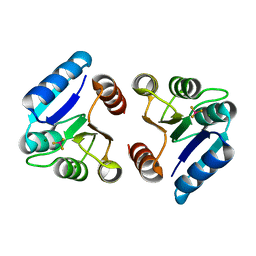 | | BeF3 Activated DrrD Receiver Domain | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, DNA BINDING RESPONSE REGULATOR D, MAGNESIUM ION | | Authors: | Robinson, V.L, Stock, A.M. | | Deposit date: | 2010-06-23 | | Release date: | 2010-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Regulation of response regulator autophosphorylation through interdomain contacts.
J.Biol.Chem., 285, 2010
|
|
3NNS
 
 | | BeF3 Activated DrrB Receiver Domain | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, DNA BINDING RESPONSE REGULATOR B, MAGNESIUM ION | | Authors: | Robinson, V.L, Stock, A.M. | | Deposit date: | 2010-06-24 | | Release date: | 2010-08-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Regulation of response regulator autophosphorylation through interdomain contacts.
J.Biol.Chem., 285, 2010
|
|
1BC5
 
 | |
4GF8
 
 | | Crystal Structure of the Chitin Oligasaccharide Binding Protein | | Descriptor: | Peptide ABC transporter, periplasmic peptide-binding protein, SULFATE ION | | Authors: | Xu, S, Li, X, Gu, L, Roseman, R, Stock, A.M. | | Deposit date: | 2012-08-03 | | Release date: | 2013-08-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Chitin catabolic cascade in the marine bacterium Vibrio cholerae: properties, structure and functions of a periplasmic chitooligosaccharide binding protein (CBP)
To be Published
|
|
3NHZ
 
 | | Structure of N-terminal Domain of MtrA | | Descriptor: | MAGNESIUM ION, Two component system transcriptional regulator mtrA | | Authors: | Barbieri, C.M, Mack, T.R, Robinson, V.L, Miller, M.T, Stock, A.M. | | Deposit date: | 2010-06-14 | | Release date: | 2010-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Regulation of response regulator autophosphorylation through interdomain contacts.
J.Biol.Chem., 285, 2010
|
|
1ZGZ
 
 | |
4GFR
 
 | | Crystal Structure of the liganded Chitin Oligasaccharide Binding Protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, MANGANESE (II) ION, Peptide ABC transporter, ... | | Authors: | Xu, S, Li, X, Gu, L, Roseman, R, Stock, A.M. | | Deposit date: | 2012-08-03 | | Release date: | 2013-08-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Chitin catabolic cascade in the marine bacterium Vibrio cholerae: properties, structure and functions of a periplasmic chitooligosaccharide binding protein (CBP)
To be Published
|
|
1ZH4
 
 | |
1ZES
 
 | | BeF3- activated PhoB receiver domain | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, Phosphate regulon transcriptional regulatory protein phoB | | Authors: | Bachhawat, P, Montelione, G.T, Stock, A.M. | | Deposit date: | 2005-04-19 | | Release date: | 2005-09-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanism of Activation for Transcription Factor PhoB Suggested by Different Modes of Dimerization in the Inactive and Active States.
Structure, 13, 2005
|
|
4GT8
 
 | |
2FMF
 
 | |
2FMK
 
 | | Crystal structure of Mg2+ and BeF3- bound CheY in complex with CheZ 200-214 solved from a P2(1)2(1)2 crystal grown in MES (pH 6.0) | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, C-terminal 15-mer from Chemotaxis protein cheZ, Chemotaxis protein cheY, ... | | Authors: | Guhaniyogi, J, Robinson, V.L, Stock, A.M. | | Deposit date: | 2006-01-09 | | Release date: | 2006-05-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Crystal Structures of Beryllium Fluoride-free and Beryllium Fluoride-bound CheY in Complex with the Conserved C-terminal Peptide of CheZ Reveal Dual Binding Modes Specific to CheY Conformation
J.Mol.Biol., 359, 2006
|
|
2FMH
 
 | | Crystal structure of Mg2+ and BeF3- bound CheY in complex with CheZ 200-214 solved from a F432 crystal grown in Tris (pH 8.4) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BERYLLIUM TRIFLUORIDE ION, C-terminal 15-mer from Chemotaxis protein cheZ, ... | | Authors: | Guhaniyogi, J, Robinson, V.L, Stock, A.M. | | Deposit date: | 2006-01-09 | | Release date: | 2006-05-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Crystal Structures of Beryllium Fluoride-free and Beryllium Fluoride-bound CheY in Complex with the Conserved C-terminal Peptide of CheZ Reveal Dual Binding Modes Specific to CheY Conformation.
J.Mol.Biol., 359, 2006
|
|
