3PAH
 
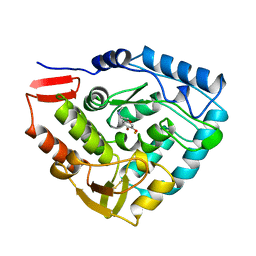 | | HUMAN PHENYLALANINE HYDROXYLASE CATALYTIC DOMAIN DIMER WITH BOUND ADRENALINE INHIBITOR | | Descriptor: | 4-[(1S)-1-hydroxy-2-(methylamino)ethyl]benzene-1,2-diol, FE (III) ION, PHENYLALANINE HYDROXYLASE | | Authors: | Erlandsen, H, Flatmark, T, Stevens, R.C. | | Deposit date: | 1998-08-20 | | Release date: | 1999-04-27 | | Last modified: | 2013-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic analysis of the human phenylalanine hydroxylase catalytic domain with bound catechol inhibitors at 2.0 A resolution.
Biochemistry, 37, 1998
|
|
3PBL
 
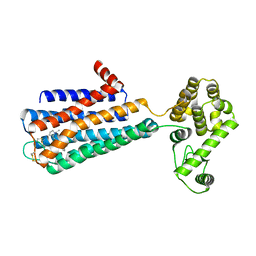 | | Structure of the human dopamine D3 receptor in complex with eticlopride | | Descriptor: | 3-chloro-5-ethyl-N-{[(2S)-1-ethylpyrrolidin-2-yl]methyl}-6-hydroxy-2-methoxybenzamide, D(3) dopamine receptor, Lysozyme chimera, ... | | Authors: | Chien, E.Y.T, Liu, W, Han, G.W, Katritch, V, Zhao, Q, Cherezov, V, Stevens, R.C, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D), GPCR Network (GPCR) | | Deposit date: | 2010-10-20 | | Release date: | 2010-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structure of the human dopamine d3 receptor in complex with a d2/d3 selective antagonist.
Science, 330, 2010
|
|
3OE6
 
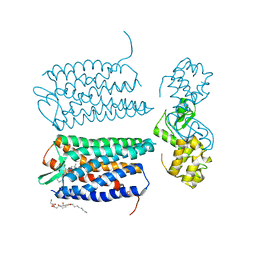 | | Crystal structure of the CXCR4 chemokine receptor in complex with a small molecule antagonist IT1t in I222 spacegroup | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (6,6-dimethyl-5,6-dihydroimidazo[2,1-b][1,3]thiazol-3-yl)methyl N,N'-dicyclohexylimidothiocarbamate, C-X-C chemokine receptor type 4, ... | | Authors: | Wu, B, Mol, C.D, Han, G.W, Katritch, V, Chien, E.Y.T, Liu, W, Cherezov, V, Stevens, R.C, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D), GPCR Network (GPCR) | | Deposit date: | 2010-08-12 | | Release date: | 2010-10-27 | | Last modified: | 2021-10-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists.
Science, 330, 2010
|
|
3OE8
 
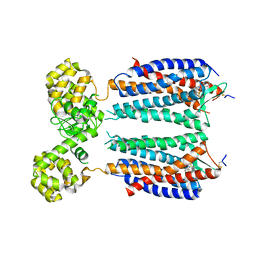 | | Crystal structure of the CXCR4 chemokine receptor in complex with a small molecule antagonist IT1t in P1 spacegroup | | Descriptor: | (6,6-dimethyl-5,6-dihydroimidazo[2,1-b][1,3]thiazol-3-yl)methyl N,N'-dicyclohexylimidothiocarbamate, C-X-C chemokine receptor type 4, Lysozyme Chimera | | Authors: | Wu, B, Mol, C.D, Han, G.W, Katritch, V, Chien, E.Y.T, Liu, W, Cherezov, V, Stevens, R.C, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D), GPCR Network (GPCR) | | Deposit date: | 2010-08-12 | | Release date: | 2010-10-27 | | Last modified: | 2021-10-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists.
Science, 330, 2010
|
|
3OJ8
 
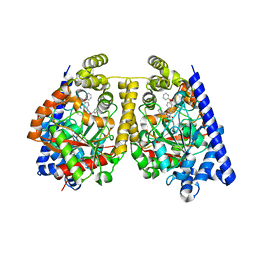 | | Alpha-Ketoheterocycle Inhibitors of Fatty Acid Amide Hydrolase Containing Additional Conformational Contraints in the Acyl Side Chain | | Descriptor: | (S)-[(2S)-6-phenoxy-1,2,3,4-tetrahydronaphthalen-2-yl](5-pyridin-2-yl-1,3-oxazol-2-yl)methanol, CHLORIDE ION, Fatty-acid amide hydrolase 1 | | Authors: | Mileni, M, Stevens, R.C, Boger, D.L. | | Deposit date: | 2010-08-20 | | Release date: | 2011-07-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | alpha-Ketoheterocycle Inhibitors of Fatty Acid Amide Hydrolase Containing Additional Conformational Contraints in the Acyl Side Chain
J.Med.Chem., 54, 2011
|
|
3PR0
 
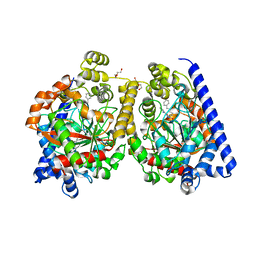 | | Crystal Structure of a Covalently Bound alpha-Ketoheterocycle Inhibitor (Phenhexyl/Oxadiazole/Pyridine) to a Humanized Variant of Fatty Acid Amide Hydrolase | | Descriptor: | 7-phenyl-1-[5-(pyridin-2-yl)-1,3,4-oxadiazol-2-yl]heptane-1,1-diol, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Mileni, M, Han, G.W, Boger, D.L, Stevens, R.C. | | Deposit date: | 2010-11-29 | | Release date: | 2011-11-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Fluoride-mediated capture of a noncovalent bound state of a reversible covalent enzyme inhibitor: X-ray crystallographic analysis of an exceptionally potent alpha-ketoheterocycle inhibitor of fatty acid amide hydrolase.
J.Am.Chem.Soc., 133, 2011
|
|
3PPM
 
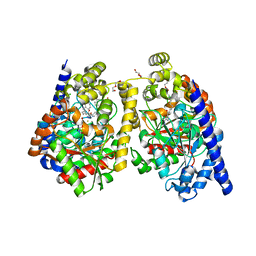 | | Crystal Structure of a Noncovalently Bound alpha-Ketoheterocycle Inhibitor (Phenhexyl/Oxadiazole/Pyridine) to a Humanized Variant of Fatty Acid Amide Hydrolase | | Descriptor: | 1-DODECANOL, 7-phenyl-1-[5-(pyridin-2-yl)-1,3,4-oxadiazol-2-yl]heptan-1-one, CHLORIDE ION, ... | | Authors: | Mileni, M, Han, G.W, Boger, D.L, Stevens, R.C. | | Deposit date: | 2010-11-24 | | Release date: | 2011-11-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Fluoride-mediated capture of a noncovalent bound state of a reversible covalent enzyme inhibitor: X-ray crystallographic analysis of an exceptionally potent alpha-ketoheterocycle inhibitor of fatty acid amide hydrolase.
J.Am.Chem.Soc., 133, 2011
|
|
3ODU
 
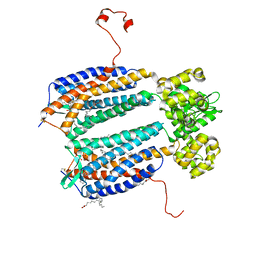 | | The 2.5 A structure of the CXCR4 chemokine receptor in complex with small molecule antagonist IT1t | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (6,6-dimethyl-5,6-dihydroimidazo[2,1-b][1,3]thiazol-3-yl)methyl N,N'-dicyclohexylimidothiocarbamate, C-X-C chemokine receptor type 4, ... | | Authors: | Wu, B, Mol, C.D, Han, G.W, Katritch, V, Chien, E.Y.T, Liu, W, Cherezov, V, Stevens, R.C, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D), GPCR Network (GPCR) | | Deposit date: | 2010-08-11 | | Release date: | 2010-10-27 | | Last modified: | 2021-10-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists.
Science, 330, 2010
|
|
3POY
 
 | | Crystal Structure of the alpha-Neurexin-1 ectodomain, LNS 2-6 | | Descriptor: | Neurexin-1-alpha, beta-D-glucopyranose, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Miller, M.T, Mileni, M, Comoletti, D, Stevens, R.C, Harel, M, Taylor, P. | | Deposit date: | 2010-11-23 | | Release date: | 2011-06-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | The crystal structure of the alpha-neurexin-1 extracellular region reveals a hinge point for mediating synaptic adhesion and function.
Structure, 19, 2011
|
|
3QIY
 
 | | Crystal Structure of BoNT/A LC complexed with Hydroxamate-based Inhibitor PT-1 | | Descriptor: | 1,2-ETHANEDIOL, 4-[bis(4-chlorobenzyl)amino]-N-hydroxybutanamide, Botulinum neurotoxin type A, ... | | Authors: | Thompson, A.A, Han, G.W, Stevens, R.C. | | Deposit date: | 2011-01-28 | | Release date: | 2011-04-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Characterization of Three Novel Hydroxamate-Based Zinc Chelating Inhibitors of the Clostridium botulinum Serotype A Neurotoxin Light Chain Metalloprotease Reveals a Compact Binding Site Resulting from 60/70 Loop Flexibility.
Biochemistry, 50, 2011
|
|
3QJ0
 
 | | Crystal Structure of BoNT/A LC complexed with Hydroxamate-based Inhibitor PT-3 | | Descriptor: | (4R)-4-(4-chlorophenoxy)-1-[(4-chlorophenyl)sulfonyl]-N-hydroxy-L-prolinamide, 1,2-ETHANEDIOL, Botulinum neurotoxin type A, ... | | Authors: | Thompson, A.A, Han, G.W, Stevens, R.C. | | Deposit date: | 2011-01-28 | | Release date: | 2011-04-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Structural Characterization of Three Novel Hydroxamate-Based Zinc Chelating Inhibitors of the Clostridium botulinum Serotype A Neurotoxin Light Chain Metalloprotease Reveals a Compact Binding Site Resulting from 60/70 Loop Flexibility.
Biochemistry, 50, 2011
|
|
3QIZ
 
 | | Crystal Structure of BoNT/A LC complexed with Hydroxamate-based Inhibitor PT-2 | | Descriptor: | (2S,4R)-2-(2-{[3-(4-fluoro-3-methylphenyl)propyl](methyl)amino}ethyl)-4-(4-fluorophenyl)-N-hydroxy-4-methoxybutanamide, Botulinum neurotoxin type A, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Thompson, A.A, Han, G.W, Stevens, R.C. | | Deposit date: | 2011-01-28 | | Release date: | 2011-04-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Characterization of Three Novel Hydroxamate-Based Zinc Chelating Inhibitors of the Clostridium botulinum Serotype A Neurotoxin Light Chain Metalloprotease Reveals a Compact Binding Site Resulting from 60/70 Loop Flexibility.
Biochemistry, 50, 2011
|
|
2IDY
 
 | | NMR Structure of the SARS-CoV non-structural protein nsp3a | | Descriptor: | NSP3 | | Authors: | Serrano, P, Almeida, M.S, Johnson, M.A, Horst, R, Herrmann, T, Joseph, J, Saikatendu, K, Subramanian, V, Stevens, R.C, Kuhn, P, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2006-09-15 | | Release date: | 2006-12-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance structure of the N-terminal domain of nonstructural protein 3 from the severe acute respiratory syndrome coronavirus.
J.Virol., 81, 2007
|
|
2GRI
 
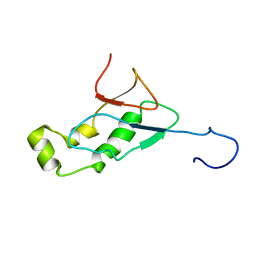 | | NMR Structure of the SARS-CoV non-structural protein nsp3a | | Descriptor: | NSP3 | | Authors: | Serrano, P, Almeida, M.S, Johnson, M.A, Herrmann, T, Saikatendu, K.S, Joseph, J, Subramanian, V, Neuman, B.W, Buchmeier, M.J, Stevens, R.C, Kuhn, P, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2006-04-24 | | Release date: | 2006-12-19 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance structure of the N-terminal domain of nonstructural protein 3 from the severe acute respiratory syndrome coronavirus.
J.Virol., 81, 2007
|
|
3QIX
 
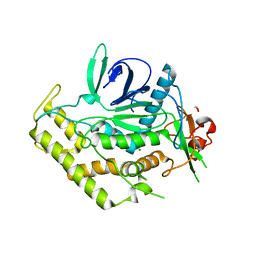 | | Crystal Structure of BoNT/A LC with Zinc bound | | Descriptor: | 1,2-ETHANEDIOL, Botulinum neurotoxin type A, ZINC ION | | Authors: | Thompson, A.A, Han, G.W, Stevens, R.C. | | Deposit date: | 2011-01-28 | | Release date: | 2011-04-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.413 Å) | | Cite: | Structural Characterization of Three Novel Hydroxamate-Based Zinc Chelating Inhibitors of the Clostridium botulinum Serotype A Neurotoxin Light Chain Metalloprotease Reveals a Compact Binding Site Resulting from 60/70 Loop Flexibility.
Biochemistry, 50, 2011
|
|
3V2W
 
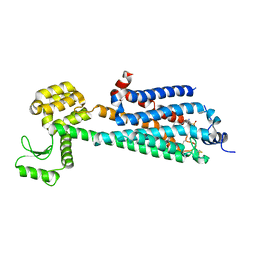 | | Crystal Structure of a Lipid G protein-Coupled Receptor at 3.35A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Sphingosine 1-phosphate receptor 1, Lysozyme chimera, ... | | Authors: | Hanson, M.A, Roth, C.B, Jo, E, Griffith, M.T, Scott, F.L, Reinhart, G, Desale, H, Clemons, B, Cahalan, S.M, Schuerer, S.C, Sanna, M.G, Han, G.W, Kuhn, P, Rosen, H, Stevens, R.C, GPCR Network (GPCR) | | Deposit date: | 2011-12-12 | | Release date: | 2012-02-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Crystal structure of a lipid G protein-coupled receptor.
Science, 335, 2012
|
|
3V2Y
 
 | | Crystal Structure of a Lipid G protein-Coupled Receptor at 2.80A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Sphingosine 1-phosphate receptor 1, Lysozyme chimera (E.C.3.2.1.17), ... | | Authors: | Hanson, M.A, Roth, C.B, Jo, E, Griffith, M.T, Scott, F.L, Reinhart, G, Desale, H, Clemons, B, Cahalan, S.M, Schuerer, S.C, Sanna, M.G, Han, G.W, Kuhn, P, Rosen, H, Stevens, R.C, GPCR Network (GPCR) | | Deposit date: | 2011-12-12 | | Release date: | 2012-02-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a lipid G protein-coupled receptor.
Science, 335, 2012
|
|
2HSX
 
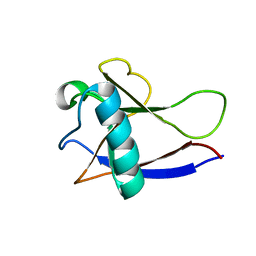 | | NMR Structure of the nonstructural protein 1 (nsp1) from the SARS coronavirus | | Descriptor: | Leader protein; p65 homolog; NSP1 (EC 3.4.22.-) | | Authors: | Almeida, M.S, Herrmann, T, Geralt, M, Johnson, M.A, Saikatendu, K, Joseph, J, Subramanian, R.C, Neuman, B.W, Buchmeier, M.J, Stevens, R.C, Kuhn, P, Wilson, I.A, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2006-07-24 | | Release date: | 2007-02-06 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Novel beta-barrel fold in the nuclear magnetic resonance structure of the replicase nonstructural protein 1 from the severe acute respiratory syndrome coronavirus.
J.Virol., 81, 2007
|
|
1AJ7
 
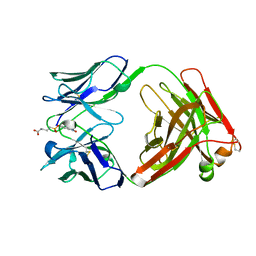 | | IMMUNOGLOBULIN 48G7 GERMLINE FAB ANTIBODY COMPLEXED WITH HAPTEN 5-(PARA-NITROPHENYL PHOSPHONATE)-PENTANOIC ACID. AFFINITY MATURATION OF AN ESTEROLYTIC ANTIBODY | | Descriptor: | 5-(PARA-NITROPHENYL PHOSPHONATE)-PENTANOIC ACID, IMMUNOGLOBULIN 48G7 FAB (HEAVY CHAIN), IMMUNOGLOBULIN 48G7 FAB (LIGHT CHAIN) | | Authors: | Wedemayer, G.J, Wang, L.H, Patten, P.A, Schultz, P.G, Stevens, R.C. | | Deposit date: | 1997-05-15 | | Release date: | 1997-11-12 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insights into the evolution of an antibody combining site.
Science, 276, 1997
|
|
1E74
 
 | | NMR SOLUTION STRUCTURE OF ALPHA-CONOTOXIN IM1 POINT MUTATION VARIANT R11E | | Descriptor: | ALPHA-CONOTOXIN IM1(R11E) | | Authors: | Rogers, J.P, Luginbuhl, P, Pemberton, K, Harty, P, Wemmer, D.E, Stevens, R.C. | | Deposit date: | 2000-08-24 | | Release date: | 2000-12-27 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure-Activity Relationships in a Peptidic Alpha7 Nicotinic Acetylcholine Receptor Antagonist
J.Mol.Biol., 304, 2000
|
|
1E76
 
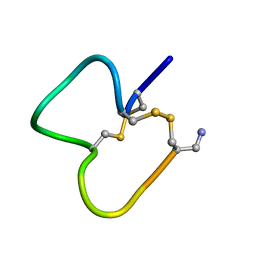 | | NMR SOLUTION STRUCTURE OF ALPHA-CONOTOXIN IM1 POINT MUTATION VARIANT D5N | | Descriptor: | ALPHA-CONOTOXIN IM1(D5N) | | Authors: | Rogers, J.P, Luginbuhl, P, Pemberton, K, Harty, P, Wemmer, D.E, Stevens, R.C. | | Deposit date: | 2000-08-24 | | Release date: | 2000-12-27 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure-Activity Relationships in a Peptidic Alpha7 Nicotinic Acetylcholine Receptor Antagonist
J.Mol.Biol., 304, 2000
|
|
1E75
 
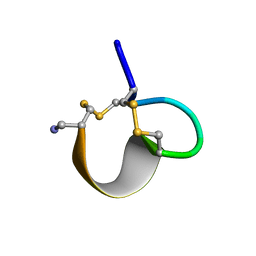 | | NMR SOLUTION STRUCTURE OF ALPHA-CONOTOXIN IM1 POINT MUTATION VARIANT R7L | | Descriptor: | ALPHA-CONOTOXIN IM1(R7L) | | Authors: | Rogers, J.P, Luginbuhl, P, Pemberton, K, Harty, P, Wemmer, D.E, Stevens, R.C. | | Deposit date: | 2000-08-24 | | Release date: | 2000-12-27 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure-Activity Relationships in a Peptidic Alpha7 Nicotinic Acetylcholine Receptor Antagonist
J.Mol.Biol., 304, 2000
|
|
3IQS
 
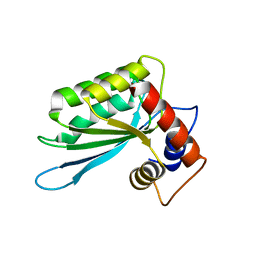 | | Crystal structure of the anti-viral APOBEC3G catalytic domain | | Descriptor: | DNA dC->dU-editing enzyme APOBEC-3G, ZINC ION | | Authors: | Holden, L.G, Prochnow, C, Chang, Y.P, Bransteitter, R, Chelico, L, Sen, U, Stevens, R.C, Goodman, R.F, Chen, X.S. | | Deposit date: | 2009-08-20 | | Release date: | 2009-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the anti-viral APOBEC3G catalytic domain and functional implications.
Nature, 456, 2008
|
|
1FL6
 
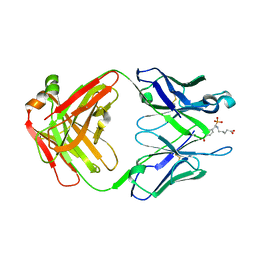 | | THE HAPTEN COMPLEXED GERMLINE PRECURSOR TO SULFIDE OXIDASE CATALYTIC ANTIBODY 28B4 | | Descriptor: | 1-[N-4'-NITROBENZYL-N-4'-CARBOXYBUTYLAMINO]METHYLPHOSPHONIC ACID, ANTIBODY GERMLINE PRECURSOR TO 28B4 | | Authors: | Yin, J, Mundorff, E.C, Yang, P.L, Wendt, K.U, Hanway, D, Stevens, R.C, Schultz, P.G. | | Deposit date: | 2000-08-11 | | Release date: | 2001-11-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A comparative analysis of the immunological evolution of antibody 28B4.
Biochemistry, 40, 2001
|
|
1FL5
 
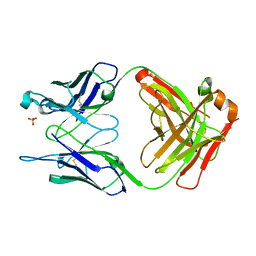 | | THE UNLIGANDED GERMLINE PRECURSOR TO THE SULFIDE OXIDASE CATALYTIC ANTIBODY 28B4. | | Descriptor: | ANTIBODY GERMLINE PRECURSOR TO ANTIBODY 28B4, SULFATE ION | | Authors: | Yin, J, Mundorff, E.C, Yang, P.L, Wendt, K.U, Hanway, D, Stevens, R.C, Schultz, P.G. | | Deposit date: | 2000-08-11 | | Release date: | 2001-11-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A comparative analysis of the immunological evolution of antibody 28B4.
Biochemistry, 40, 2001
|
|
