1QQ1
 
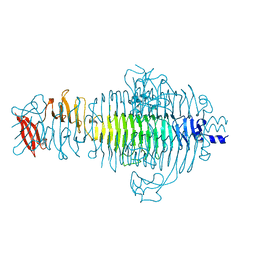 | | TAILSPIKE PROTEIN, MUTANT E359G | | Descriptor: | TAILSPIKE PROTEIN | | Authors: | Schuler, B, Furst, F, Osterroth, F, Steinbacher, S, Huber, R, Seckler, R. | | Deposit date: | 1999-06-10 | | Release date: | 2000-04-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Plasticity and steric strain in a parallel beta-helix: rational mutations in the P22 tailspike protein.
Proteins, 39, 2000
|
|
1U7Z
 
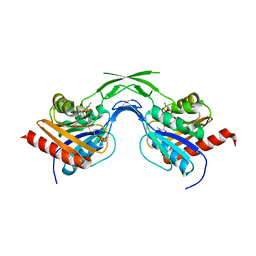 | | Phosphopantothenoylcysteine synthetase from E. coli, 4'-phosphopantothenoyl-CMP complex | | Descriptor: | Coenzyme A biosynthesis bifunctional protein coaBC, PHOSPHORIC ACID MONO-[3-(3-{[5-(4-AMINO-2-OXO-2H-PYRIMIDIN-1-YL)-3,4- DIHYDROXY-TETRAHYDRO-FURAN-2- YLMETHOXY]-HYDROXY-PHOSPHORYLOXY}-3-OXO-PROPYLCARBAMOYL)-3-HYDROXY-2,2- DIMETHYL-PROPYL] ESTER | | Authors: | Stanitzek, S, Augustin, M.A, Huber, R, Kupke, T, Steinbacher, S. | | Deposit date: | 2004-08-04 | | Release date: | 2004-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of CTP-Dependent Peptide Bond Formation in Coenzyme A Biosynthesis Catalyzed by Escherichia coli PPC Synthetase
STRUCTURE, 12, 2004
|
|
1U7U
 
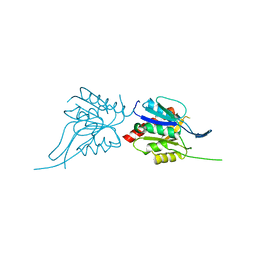 | | Phosphopantothenoylcysteine synthetase from E. coli | | Descriptor: | Coenzyme A biosynthesis bifunctional protein coaBC | | Authors: | Stanitzek, S, Augustin, M.A, Huber, R, Kupke, T, Steinbacher, S. | | Deposit date: | 2004-08-04 | | Release date: | 2004-11-30 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis of CTP-Dependent Peptide Bond Formation in Coenzyme A Biosynthesis Catalyzed by Escherichia coli PPC Synthetase
STRUCTURE, 12, 2004
|
|
1U80
 
 | | Phosphopantothenoylcysteine synthetase from E. coli, CMP complex | | Descriptor: | CYTIDINE-5'-MONOPHOSPHATE, Coenzyme A biosynthesis bifunctional protein coaBC, PHOSPHATE ION | | Authors: | Stanitzek, S, Augustin, M.A, Huber, R, Kupke, T, Steinbacher, S. | | Deposit date: | 2004-08-04 | | Release date: | 2004-11-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural Basis of CTP-Dependent Peptide Bond Formation in Coenzyme A Biosynthesis Catalyzed by Escherichia coli PPC Synthetase
STRUCTURE, 12, 2004
|
|
1U7W
 
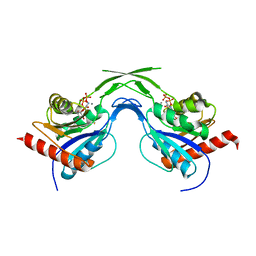 | | Phosphopantothenoylcysteine synthetase from E. coli, CTP-complex | | Descriptor: | CALCIUM ION, CYTIDINE-5'-TRIPHOSPHATE, Coenzyme A biosynthesis bifunctional protein coaBC | | Authors: | Stanitzek, S, Augustin, M.A, Huber, R, Kupke, T, Steinbacher, S. | | Deposit date: | 2004-08-04 | | Release date: | 2004-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis of CTP-Dependent Peptide Bond Formation in Coenzyme A Biosynthesis Catalyzed by Escherichia coli PPC Synthetase
STRUCTURE, 12, 2004
|
|
1N05
 
 | | Crystal Structure of Schizosaccharomyces pombe Riboflavin Kinase Reveals a Novel ATP and Riboflavin Binding Fold | | Descriptor: | putative Riboflavin kinase | | Authors: | Bauer, S, Kemter, K, Bacher, A, Huber, R, Fischer, M, Steinbacher, S. | | Deposit date: | 2002-10-11 | | Release date: | 2003-02-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Schizosaccharomyces pombe Riboflavin Kinase Reveals a Novel ATP and Riboflavin Binding Fold
J.Mol.Biol., 326, 2003
|
|
6AZW
 
 | | IDO1/FXB-001116 crystal structure | | Descriptor: | (2R)-N-(4-cyanophenyl)-2-[cis-4-(quinolin-4-yl)cyclohexyl]propanamide, Indoleamine 2,3-dioxygenase 1 | | Authors: | Lewis, H.A, Lammens, A, Steinbacher, S. | | Deposit date: | 2017-09-13 | | Release date: | 2018-03-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Immune-modulating enzyme indoleamine 2,3-dioxygenase is effectively inhibited by targeting its apo-form.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
1N06
 
 | | Crystal Structure of Schizosaccharomyces pombe Riboflavin Kinase Reveals a Novel ATP and Riboflavin Binding Fold | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, PUTATIVE riboflavin kinase | | Authors: | Bauer, S, Kemter, K, Bacher, A, Huber, R, Fischer, M, Steinbacher, S. | | Deposit date: | 2002-10-11 | | Release date: | 2003-02-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Schizosaccharomyces pombe Riboflavin Kinase Reveals a Novel ATP and Riboflavin Binding Fold
J.Mol.Biol., 326, 2003
|
|
1N08
 
 | | Crystal Structure of Schizosaccharomyces pombe Riboflavin Kinase Reveals a Novel ATP and Riboflavin Binding Fold | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ZINC ION, putative riboflavin kinase | | Authors: | Bauer, S, Kemter, K, Bacher, A, Huber, R, Fischer, M, Steinbacher, S. | | Deposit date: | 2002-10-11 | | Release date: | 2003-02-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of Schizosaccharomyces pombe Riboflavin Kinase Reveals a Novel ATP and Riboflavin Binding Fold
J.Mol.Biol., 326, 2003
|
|
1N07
 
 | | Crystal Structure of Schizosaccharomyces pombe Riboflavin Kinase Reveals a Novel ATP and Riboflavin Binding Fold | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, FLAVIN MONONUCLEOTIDE, PUTATIVE riboflavin kinase | | Authors: | Bauer, S, Kemter, K, Bacher, A, Huber, R, Fischer, M, Steinbacher, S. | | Deposit date: | 2002-10-11 | | Release date: | 2003-02-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal Structure of Schizosaccharomyces pombe Riboflavin Kinase Reveals a Novel ATP and Riboflavin Binding Fold
J.Mol.Biol., 326, 2003
|
|
4L3P
 
 | | Crystal Structure of 2-(1-benzothiophen-7-yl)-4-[1-(piperidin-4-yl)-1H-pyrazol-4-yl]furo[2,3-c]pyridin-7-amine bound to TAK1-TAB1 | | Descriptor: | 2-(1-benzothiophen-7-yl)-4-[1-(piperidin-4-yl)-1H-pyrazol-4-yl]furo[2,3-c]pyridin-7-amine, Mitogen-activated protein kinase kinase kinase 7, TGF-beta-activated kinase 1 and MAP3K7-binding protein 1 chimera | | Authors: | Wang, J, Hornberger, K.R, Crew, A.P, Steinbacher, S, Maskos, K, Moertl, M. | | Deposit date: | 2013-06-06 | | Release date: | 2013-06-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Discovery and optimization of 7-aminofuro[2,3-c]pyridine inhibitors of TAK1.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
1SP8
 
 | | 4-Hydroxyphenylpyruvate Dioxygenase | | Descriptor: | 4-Hydroxyphenylpyruvate Dioxygenase, FE (II) ION | | Authors: | Fritze, I.M, Linden, L, Freigang, J, Auerbach, G, Huber, R, Steinbacher, S. | | Deposit date: | 2004-03-16 | | Release date: | 2004-09-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structures of Zea mays and Arabidopsis 4-Hydroxyphenylpyruvate Dioxygenase
Plant physiol., 134, 2004
|
|
1SP9
 
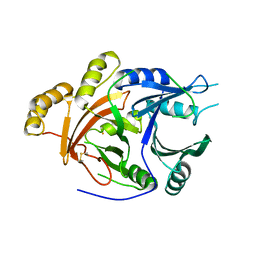 | | 4-Hydroxyphenylpyruvate Dioxygenase | | Descriptor: | 4-hydroxyphenylpyruvate dioxygenase, FE (II) ION | | Authors: | Fritze, I.M, Linden, L, Freigang, J, Auerbach, G, Huber, R, Steinbacher, S. | | Deposit date: | 2004-03-16 | | Release date: | 2004-09-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The crystal structures of Zea mays and Arabidopsis 4-Hydroxyphenylpyruvate Dioxygenase
Plant Physiol., 134, 2004
|
|
1P3Y
 
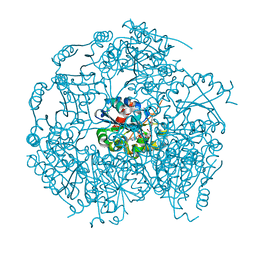 | | MrsD from Bacillus sp. HIL-Y85/54728 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, MrsD protein | | Authors: | Blaesse, M, Kupke, T, Huber, R, Steinbacher, S. | | Deposit date: | 2003-04-19 | | Release date: | 2003-08-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Structure of MrsD, an FAD-binding protein of the HFCD family.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1QA2
 
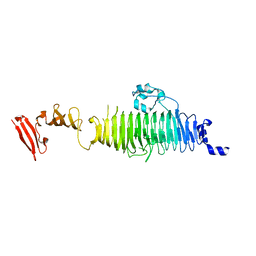 | | TAILSPIKE PROTEIN, MUTANT A334V | | Descriptor: | TAILSPIKE PROTEIN | | Authors: | Baxa, U, Steinbacher, S, Weintraub, A, Huber, R, Seckler, R. | | Deposit date: | 1999-04-10 | | Release date: | 2000-01-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mutations improving the folding of phage P22 tailspike protein affect its receptor binding activity.
J.Mol.Biol., 293, 1999
|
|
1QA1
 
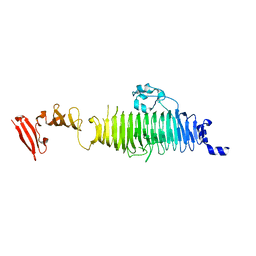 | | TAILSPIKE PROTEIN, MUTANT V331G | | Descriptor: | TAILSPIKE PROTEIN | | Authors: | Baxa, U, Steinbacher, S, Weintraub, A, Huber, R, Seckler, R. | | Deposit date: | 1999-04-10 | | Release date: | 2000-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mutations improving the folding of phage P22 tailspike protein affect its receptor binding activity.
J.Mol.Biol., 293, 1999
|
|
1QA3
 
 | | TAILSPIKE PROTEIN, MUTANT A334I | | Descriptor: | TAILSPIKE PROTEIN | | Authors: | Baxa, U, Steinbacher, S, Weintraub, A, Huber, R, Seckler, R. | | Deposit date: | 1999-04-10 | | Release date: | 2000-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mutations improving the folding of phage P22 tailspike protein affect its receptor binding activity.
J.Mol.Biol., 293, 1999
|
|
3LXG
 
 | | Crystal structure of rat phosphodiesterase 10A in complex with ligand WEB-3 | | Descriptor: | 2-methoxy-6,7-dimethyl-9-propylimidazo[1,5-a]pyrido[3,2-e]pyrazine, MAGNESIUM ION, ZINC ION, ... | | Authors: | Mosbacher, T, Jestel, A, Steinbacher, S. | | Deposit date: | 2010-02-25 | | Release date: | 2010-05-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of imidazo[1,5-a]pyrido[3,2-e]pyrazines as a new class of phosphodiesterase 10A inhibitiors.
J.Med.Chem., 53, 2010
|
|
4GV1
 
 | | PKB alpha in complex with AZD5363 | | Descriptor: | 4-amino-N-[(1S)-1-(4-chlorophenyl)-3-hydroxypropyl]-1-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)piperidine-4-carboxamide, GLYCEROL, RAC-alpha serine/threonine-protein kinase | | Authors: | Addie, M, Ballard, P, Bird, G, Buttar, D, Currie, G, Davies, B, Debreczeni, J, Dry, H, Dudley, P, Greenwood, R, Hatter, G, Jestel, A, Johnson, P.D, Kettle, J.G, Lane, C, Lamont, G, Leach, A, Luke, R.W.A, Ogilvie, D, Page, K, Pass, M, Steinbacher, S, Steuber, H, Pearson, S, Ruston, L. | | Deposit date: | 2012-08-30 | | Release date: | 2013-02-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Discovery of 4-Amino-N-[(1S)-1-(4-chlorophenyl)-3-hydroxypropyl]-1-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)piperidine-4-carboxamide (AZD5363), an Orally Bioavailable, Potent Inhibitor of Akt Kinases.
J.Med.Chem., 56, 2013
|
|
