7K9Y
 
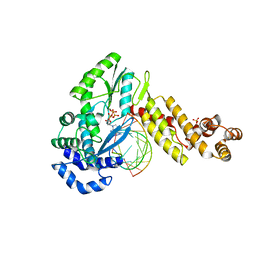 | | GsI-IIC RT Template-Switching Complex (twinned) | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, DNA (5'-D(*CP*TP*CP*CP*AP*GP*GP*CP*AP*AP*C)-3'), MAGNESIUM ION, ... | | Authors: | Stamos, J.L, Lentzsch, A.M. | | Deposit date: | 2020-09-29 | | Release date: | 2021-08-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for template switching by a group II intron-encoded non-LTR-retroelement reverse transcriptase.
J.Biol.Chem., 297, 2021
|
|
6AR5
 
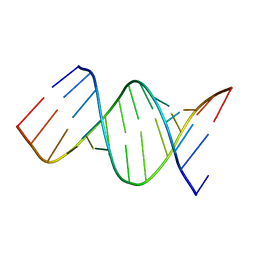 | |
6AR1
 
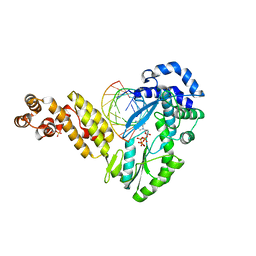 | | Structure of a Thermostable Group II Intron Reverse Transcriptase with Template-Primer and Its Functional and Evolutionary Implications (RT/Duplex (Nat)) | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, DNA, GsI-IIC RT, ... | | Authors: | Stamos, J.L, Lentzsch, A.M, Lambowitz, A.M. | | Deposit date: | 2017-08-21 | | Release date: | 2017-11-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structure of a Thermostable Group II Intron Reverse Transcriptase with Template-Primer and Its Functional and Evolutionary Implications.
Mol. Cell, 68, 2017
|
|
6AR3
 
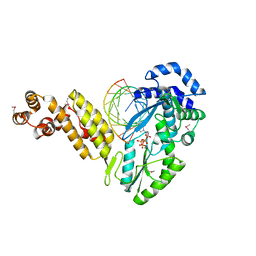 | | Structure of a Thermostable Group II Intron Reverse Transcriptase with Template-Primer and Its Functional and Evolutionary Implications (RT/Duplex (Se-Met)) | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, DNA, GsI-IIC RT, ... | | Authors: | Stamos, J.L, Lentzsch, A.M, Lambowitz, A.M. | | Deposit date: | 2017-08-21 | | Release date: | 2017-11-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.41 Å) | | Cite: | Structure of a Thermostable Group II Intron Reverse Transcriptase with Template-Primer and Its Functional and Evolutionary Implications.
Mol. Cell, 68, 2017
|
|
6DD5
 
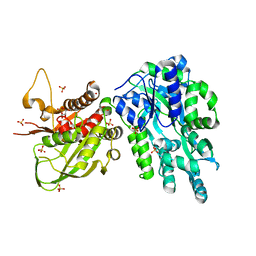 | | Crystal Structure of the Cas6 Domain of Marinomonas mediterranea MMB-1 Cas6-RT-Cas1 Fusion Protein | | Descriptor: | GLYCEROL, MMB-1 Cas6 Fused to Maltose Binding Protein,CRISPR-associated endonuclease Cas1, SULFATE ION, ... | | Authors: | Stamos, J.L, Mohr, G, Silas, S, Makarova, K.S, Markham, L.M, Yao, J, Lucas-Elio, P, Sanchez-Amat, A, Fire, A.Z, Koonin, E.V, Lambowitz, A.M. | | Deposit date: | 2018-05-09 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | A Reverse Transcriptase-Cas1 Fusion Protein Contains a Cas6 Domain Required for Both CRISPR RNA Biogenesis and RNA Spacer Acquisition.
Mol. Cell, 72, 2018
|
|
4NM5
 
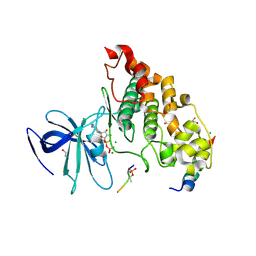 | | Crystal structure of GSK-3/Axin complex bound to phosphorylated Wnt receptor LRP6 c-motif | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Axin-1, CHLORIDE ION, ... | | Authors: | Stamos, J.L, Chu, M.L.-H, Enos, M.D, Shah, N, Weis, W.I. | | Deposit date: | 2013-11-14 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of GSK-3 inhibition by N-terminal phosphorylation and by the Wnt receptor LRP6.
Elife, 3, 2014
|
|
4NM7
 
 | | Crystal structure of GSK-3/Axin complex bound to phosphorylated Wnt receptor LRP6 e-motif | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, ADENOSINE-5'-DIPHOSPHATE, Axin-1, ... | | Authors: | Stamos, J.L, Chu, M.L.-H, Enos, M.D, Shah, N, Weis, W.I. | | Deposit date: | 2013-11-14 | | Release date: | 2014-03-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of GSK-3 inhibition by N-terminal phosphorylation and by the Wnt receptor LRP6.
Elife, 3, 2014
|
|
1M17
 
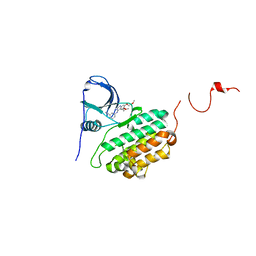 | |
1M14
 
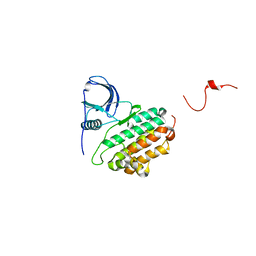 | |
1SHY
 
 | |
1RPQ
 
 | | High Affinity IgE Receptor (alpha chain) Complexed with Tight-Binding E131 'zeta' Peptide from Phage Display | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Stamos, J, Eigenbrot, C, Nakamura, G.R, Reynolds, M.E, Yin, J.P, Lowman, H.B, Fairbrother, W.J, Starovasnik, M.A. | | Deposit date: | 2003-12-03 | | Release date: | 2004-07-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Convergent Recognition of the IgE Binding Site on the High-Affinity IgE Receptor.
Structure, 12, 2004
|
|
6U9C
 
 | | The 2.2 A Crystal Structure of the Type B Chloramphenicol Acetyltransferase from Vibrio cholerae in the complex with Acetyl CoA | | Descriptor: | ACETYL COENZYME *A, CHLORIDE ION, CITRIC ACID, ... | | Authors: | Kim, Y, Maltseva, N, Stam, J, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-09-07 | | Release date: | 2019-09-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The 2.2 A Crystal Structure of the Type B Chloramphenicol Acetyltransferase from Vibrio cholerae in the complex with Acetyl CoA
To Be Published
|
|
4ZO4
 
 | | Dephospho-CoA kinase from Campylobacter jejuni. | | Descriptor: | BETA-MERCAPTOETHANOL, Dephospho-CoA kinase | | Authors: | Osipiuk, J, Zhou, M, Stam, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-05-06 | | Release date: | 2015-05-13 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Dephospho-CoA kinase from Campylobacter jejuni.
to be published
|
|
4EP1
 
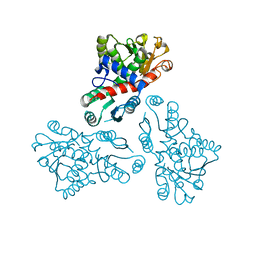 | | Crystal structure of anabolic ornithine carbamoyltransferase from Bacillus anthracis | | Descriptor: | Ornithine carbamoyltransferase | | Authors: | Shabalin, I.G, Mikolajczak, K, Stam, J, Winsor, J, Shuvalova, L, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-04-16 | | Release date: | 2012-04-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Crystal structures of anabolic ornithine carbamoyltransferase from Bacillus anthracis
To be Published
|
|
4K2H
 
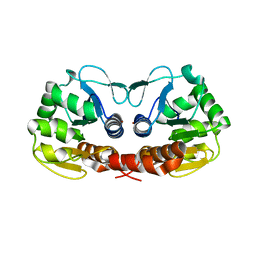 | | Crystal structure of C103A mutant of DJ-1 superfamily protein STM1931 from Salmonella typhimurium | | Descriptor: | Intracellular protease/amidase, ZINC ION | | Authors: | Shumilin, I.A, Niedzialkowska, E, Domagalski, M.J, Stam, J, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-04-09 | | Release date: | 2013-04-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of C103A mutant of DJ-1 superfamily protein STM1931 from Salmonella typhimurium
TO BE PUBLISHED
|
|
6PUB
 
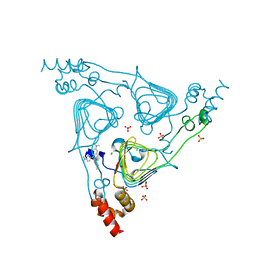 | | Crystal Structure of the Type B Chloramphenicol Acetyltransferase from Vibrio cholerae in the Complex with Crystal Violet | | Descriptor: | CHLORIDE ION, CRYSTAL VIOLET, Chloramphenicol acetyltransferase, ... | | Authors: | Kim, Y, Maltseva, N, Kuhn, M, Stam, J, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-07-18 | | Release date: | 2019-09-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Crystal Structure of the Type B Chloramphenicol Acetyltransferase from Vibrio cholerae in the Complex with Crystal Violet
To Be Published
|
|
5UJS
 
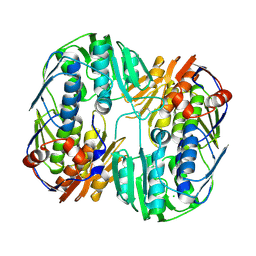 | | 2.45 Angstrom Resolution Crystal Structure of UDP-N-acetylglucosamine 1-carboxyvinyltransferase from Campylobacter jejuni. | | Descriptor: | CHLORIDE ION, UDP-N-acetylglucosamine 1-carboxyvinyltransferase | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Stam, J, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-01-18 | | Release date: | 2017-02-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | 2.45 Angstrom Resolution Crystal Structure of UDP-N-acetylglucosamine 1-carboxyvinyltransferase from Campylobacter jejuni.
To Be Published
|
|
5US8
 
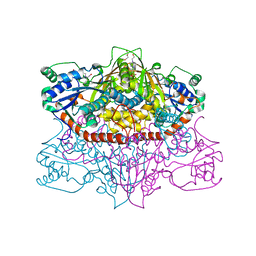 | | 2.15 Angstrom Resolution Crystal Structure of Argininosuccinate Synthase from Bordetella pertussis | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ADENOSINE, Argininosuccinate synthase, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Stam, J, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-13 | | Release date: | 2017-03-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | 2.15 Angstrom Resolution Crystal Structure of Argininosuccinate Synthase from Bordetella pertussis
To Be Published
|
|
5UTU
 
 | | 2.65 Angstrom Resolution Crystal Structure of S-adenosylhomocysteinase from Cryptosporidium parvum in Complex with SAH and NAD | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, ADENOSINE, Adenosylhomocysteinase, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Stam, J, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-15 | | Release date: | 2017-03-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | 2.65 Angstrom Resolution Crystal Structure of S-adenosylhomocysteinase from Cryptosporidium parvum in Complex with SAH and NAD
To Be Published
|
|
5DUL
 
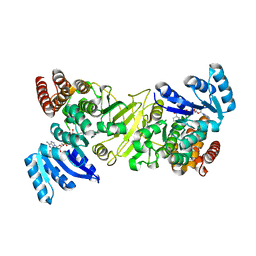 | | 1-deoxy-D-xylulose 5-phosphate reductoisomerase from Yersinia pestis in complex with NADPH | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Osipiuk, J, Mulligan, R, Stam, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-09-18 | | Release date: | 2015-09-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase from Yersinia pestis in complex with NADPH .
to be published
|
|
5EVC
 
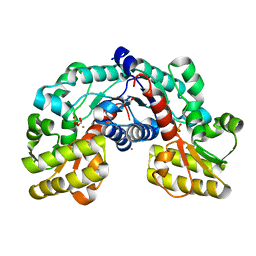 | | Crystal structure of putative aspartate racemase from Salmonella Typhimurium complexed with sulfate and potassium | | Descriptor: | CHLORIDE ION, FLUORIDE ION, FORMIC ACID, ... | | Authors: | Maltseva, N, Kim, Y, Stam, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-11-19 | | Release date: | 2015-12-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of putative aspartate racemase from Salmonella Typhimurium complexed with sulfate and potassium
To be published
|
|
5F4C
 
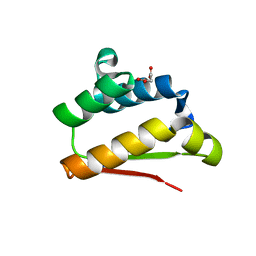 | | Crystal Structure of Ribonuclease Inhibitor Barstar from Salmonella Typhimurium | | Descriptor: | MALONATE ION, Putative cytoplasmic protein | | Authors: | Maltseva, N, Kim, Y, Mulligan, R, Stam, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-12-03 | | Release date: | 2015-12-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of Ribonuclease Inhibitor Barstar from Salmonella Typhimurium.
To Be Published
|
|
4NVR
 
 | | 2.22 Angstrom Resolution Crystal Structure of a Putative Acyltransferase from Salmonella enterica | | Descriptor: | CALCIUM ION, CHLORIDE ION, Putative acyltransferase | | Authors: | Minasov, G, Wawrzak, Z, Skarina, T, Gordon, E, Stam, J, Kwon, K, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-12-05 | | Release date: | 2013-12-18 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | 2.22 Angstrom Resolution Crystal Structure of a Putative Acyltransferase from Salmonella enterica.
TO BE PUBLISHED
|
|
4NF2
 
 | | Crystal structure of anabolic ornithine carbamoyltransferase from Bacillus anthracis in complex with carbamoyl phosphate and L-norvaline | | Descriptor: | CHLORIDE ION, NORVALINE, Ornithine carbamoyltransferase, ... | | Authors: | Shabalin, I.G, Handing, K, Cymborowski, M.T, Stam, J, Winsor, J, Shuvalova, L, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-10-30 | | Release date: | 2013-11-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal structures and kinetic properties of anabolic ornithine carbamoyltransferase from human pathogens Vibrio vulnificus and Bacillus anthracis
To be Published
|
|
4R9O
 
 | | Crystal Structure of Putative Aldo/Keto Reductase from Salmonella enterica | | Descriptor: | Putative aldo/keto reductase | | Authors: | Kim, Y, Maltseva, N, Stam, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-09-05 | | Release date: | 2014-09-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.949 Å) | | Cite: | Crystal Structure of Putative Aldo/Keto Reductase from Salmonella enterica
To be Published
|
|
