1YY1
 
 | |
1YL8
 
 | | 3D Solution Structure of [Tyr3]Octreotate derivatives in DMSO | | Descriptor: | [Tyr3]Octreotate peptide | | Authors: | Spyroulias, G.A, Galanis, A.S, Petrou, C, Vahliotis, D, Sotiriou, P, Nikolopoulou, A, Nock, B, Maina, T, Cordopatis, P. | | Deposit date: | 2005-01-19 | | Release date: | 2005-09-20 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | 3D solution structure of [Tyr3]octreotate derivatives in DMSO: structure differentiation of peptide core due to chelate group attachment and biologically active conformation.
Med.Chem., 1, 2005
|
|
1YL9
 
 | | 3D Solution Structure of [Tyr3]Octreotate derivatives in DMSO | | Descriptor: | 3-[(2-AMINOETHYL)AMINO]-2-{[(2-AMINOETHYL)AMINO]METHYL}PROPANAL, [Tyr3]Octreotate | | Authors: | Spyroulias, G.A, Galanis, A.S, Petrou, C, Vahliotis, D, Sotiriou, P, Nikolopoulou, A, Nock, B, Maina, T, Cordopatis, P. | | Deposit date: | 2005-01-19 | | Release date: | 2005-09-20 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | 3D solution structure of [Tyr3]octreotate derivatives in DMSO: structure differentiation of peptide core due to chelate group attachment and biologically active conformation.
Med.Chem., 1, 2005
|
|
1YY2
 
 | |
1GOE
 
 | |
1GO9
 
 | |
1N9V
 
 | | Differences and Similarities in Solution Structures of Angiotensin I & II: Implication for Structure-Function Relationship. | | Descriptor: | Angiotensin II | | Authors: | Spyroulias, G.A, Nikolakopoulou, P, Tzakos, A, Gerothanassis, I.P, Magafa, V, Manessi-Zoupa, E, Cordopatis, P. | | Deposit date: | 2002-11-26 | | Release date: | 2003-07-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Comparison of the solution structures of angiotensin I & II. Implication for structure-function relationship.
Eur.J.Biochem., 270, 2003
|
|
1N9U
 
 | | Differences and Similarities in Solution Structures of Angiotensin I & II: Implication for Structure-Function Relationship | | Descriptor: | Angiotensin I | | Authors: | Spyroulias, G.A, Nikolakopoulou, P, Tzakos, A, Gerothanassis, I.P, Magafa, V, Manessi-Zoupa, E, Cordopatis, P. | | Deposit date: | 2002-11-26 | | Release date: | 2003-07-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Comparison of the solution structures of angiotensin I & II. Implication for structure-function relationship.
Eur.J.Biochem., 270, 2003
|
|
5MQX
 
 | | NMR solution structure of macro domain from Venezuelan equine encephalitis virus(VEEV) in complex with ADP-ribose | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Non-structural protein3 | | Authors: | Makrynitsa, G.I, Ntonti, D, Marousis, K.D, Matsoukas, M.T, Papageorgiou, N, Coutard, B, Bentrop, D, Spyroulias, G.A. | | Deposit date: | 2016-12-21 | | Release date: | 2018-07-04 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Conformational plasticity of the VEEV macro domain is important for binding of ADP-ribose.
J.Struct.Biol., 206, 2019
|
|
2GCF
 
 | | Solution structure of the N-terminal domain of the coppper(I) ATPase PacS in its apo form | | Descriptor: | Cation-transporting ATPase pacS | | Authors: | Banci, L, Bertini, I, Ciofi-Baffoni, S, Kandias, N.G, Spyroulias, G.A, Robinson, N.J, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2006-03-14 | | Release date: | 2006-05-30 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The delivery of copper for thylakoid import observed by NMR.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2GIW
 
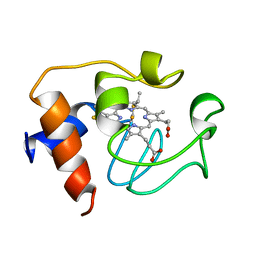 | | SOLUTION STRUCTURE OF REDUCED HORSE HEART CYTOCHROME C, NMR, 40 STRUCTURES | | Descriptor: | CYTOCHROME C, HEME C | | Authors: | Banci, L, Bertini, I, Huber, J.G, Spyroulias, G.A, Turano, P. | | Deposit date: | 1998-06-25 | | Release date: | 1998-12-09 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Solution structure of reduced horse heart cytochrome c.
J.Biol.Inorg.Chem., 4, 1999
|
|
5IQ5
 
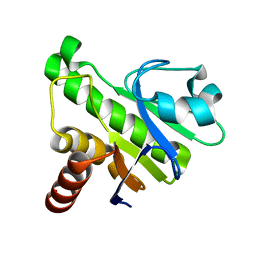 | | NMR solution structure of Mayaro virus macro domain | | Descriptor: | Macro domain | | Authors: | Melekis, E, Tsika, A.C, Bentrop, D, Papageorgiou, N, Coutard, B, Spyroulias, G.A. | | Deposit date: | 2016-03-10 | | Release date: | 2017-12-20 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Deciphering the Nucleotide and RNA Binding Selectivity of the Mayaro Virus Macro Domain.
J.Mol.Biol., 431, 2019
|
|
5ISN
 
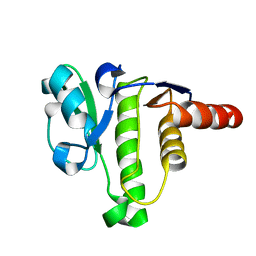 | | NMR solution structure of macro domain from Venezuelan equine encephalitis virus | | Descriptor: | Non-structural polyprotein | | Authors: | Makrynitsa, G.I, Ntonti, D, Marousis, K.D, Tsika, A.C, Papageorgiou, N, Coutard, B, Bentrop, D, Spyroulias, G.A. | | Deposit date: | 2016-03-15 | | Release date: | 2017-11-29 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Conformational plasticity of the VEEV macro domain is important for binding of ADP-ribose.
J.Struct.Biol., 206, 2019
|
|
6XXU
 
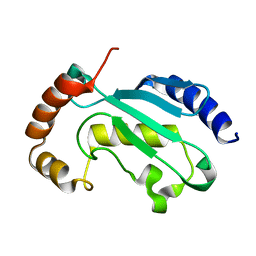 | | Solution NMR structure of the native form of UbcH7 (UBE2L3) | | Descriptor: | Ubiquitin-conjugating enzyme E2 L3 | | Authors: | Marousis, K.D, Seliami, A, Birkou, M, Episkopou, V, Spyroulias, G.A. | | Deposit date: | 2020-01-28 | | Release date: | 2020-02-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | 1H,13C,15N backbone and side-chain resonance assignment of the native form of UbcH7 (UBE2L3) through solution NMR spectroscopy.
Biomol.Nmr Assign., 14, 2020
|
|
7P2K
 
 | | Solution NMR Structure of Arginine to Cysteine mutant of Arkadia RING domain. | | Descriptor: | E3 ubiquitin-protein ligase Arkadia, ZINC ION | | Authors: | Raptis, V, Marousis, K.D, Birkou, M, Bentrop, D, Episkopou, V, Spyroulias, G.A. | | Deposit date: | 2021-07-06 | | Release date: | 2022-03-23 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Impact of a Single Nucleotide Polymorphism on the 3D Protein Structure and Ubiquitination Activity of E3 Ubiquitin Ligase Arkadia.
Front Mol Biosci, 9, 2022
|
|
5LG0
 
 | | Solution NMR structure of Tryptophan to Alanine mutant of Arkadia RING domain. | | Descriptor: | E3 ubiquitin-protein ligase Arkadia, ZINC ION | | Authors: | Birkou, M, Chasapis, C.T, Loutsidou, A.K, Bentrop, D, Lelli, M, Herrmann, T, Episkopou, V, Spyroulias, G.A. | | Deposit date: | 2016-07-05 | | Release date: | 2017-06-28 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | A Residue Specific Insight into the Arkadia E3 Ubiquitin Ligase Activity and Conformational Plasticity.
J. Mol. Biol., 429, 2017
|
|
5LG7
 
 | | Solution NMR structure of Tryptophan to Arginine mutant of Arkadia RING domain | | Descriptor: | E3 ubiquitin-protein ligase Arkadia, ZINC ION | | Authors: | Birkou, M, Chasapis, C.T, Loutsidou, A.K, Bentrop, D, Lelli, M, Herrmann, T, Episkopou, V, Spyroulias, G.A. | | Deposit date: | 2016-07-06 | | Release date: | 2017-06-28 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | A Residue Specific Insight into the Arkadia E3 Ubiquitin Ligase Activity and Conformational Plasticity.
J. Mol. Biol., 429, 2017
|
|
2K6Z
 
 | | Solution structures of copper loaded form PCuA (trans conformation of the peptide bond involving the nitrogen of P14) | | Descriptor: | COPPER (I) ION, Putative uncharacterized protein TTHA1943 | | Authors: | Abriata, L.A, Banci, L, Bertini, I, Ciofi-Baffoni, S, Gkazonis, P.A, Spyroulias, G.A, Vila, A.J, Wang, S. | | Deposit date: | 2008-07-28 | | Release date: | 2008-09-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Mechanism of Cu(A) assembly.
Nat.Chem.Biol., 4, 2008
|
|
2K70
 
 | | Solution structures of copper loaded form PCuA (cis conformation of the peptide bond involving the nitrogen of P14) | | Descriptor: | COPPER (I) ION, Putative uncharacterized protein TTHA1943 | | Authors: | Abriata, L.A, Banci, L, Bertini, I, Ciofi-Baffoni, S, Gkazonis, P.A, Spyroulias, G.A, Vila, A.J, Wang, S. | | Deposit date: | 2008-07-29 | | Release date: | 2008-09-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Mechanism of Cu(A) assembly.
Nat.Chem.Biol., 4, 2008
|
|
2K6W
 
 | | Solution structures of apo PCuA (trans conformation of the peptide bond involving the nitrogen of P14) | | Descriptor: | Putative uncharacterized protein TTHA1943 | | Authors: | Abriata, L.A, Banci, L, Bertini, I, Ciofi-Baffoni, S, Gkazonis, P, Spyroulias, G.A, Vila, A.J, Wang, S. | | Deposit date: | 2008-07-28 | | Release date: | 2008-09-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Mechanism of Cu(A) assembly.
Nat.Chem.Biol., 4, 2008
|
|
2L0R
 
 | | Conformational Dynamics of the Anthrax Lethal Factor Catalytic Center | | Descriptor: | Lethal factor | | Authors: | Dalkas, G.A, Chasapis, C.T, Gkazonis, P.V, Bentrop, D.A, Spyroulias, G.A. | | Deposit date: | 2010-07-15 | | Release date: | 2010-12-22 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Conformational dynamics of the anthrax lethal factor catalytic center.
Biochemistry, 49, 2010
|
|
2M5W
 
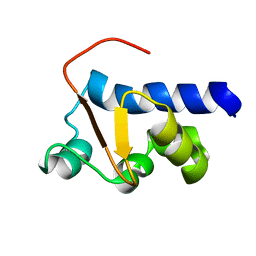 | | NMR Solution Structure of the La motif (N-terminal Domain, NTD) of Dictyostelium discoideum La protein | | Descriptor: | Lupus La protein | | Authors: | Vourtsis, D.J, Chasapis, C.T, Apostolidi, M, Stathopoulos, C, Bentrop, D, Spyroulias, G.A. | | Deposit date: | 2013-03-11 | | Release date: | 2014-03-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of the La motif (N-terminal Domain, NTD) of Dictyostelium discoideum La protein
To be Published
|
|
2KIZ
 
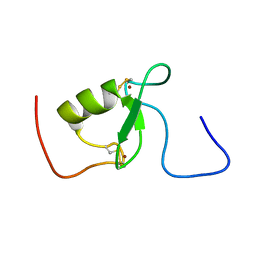 | | Solution structure of Arkadia RING-H2 finger domain | | Descriptor: | E3 ubiquitin-protein ligase Arkadia, ZINC ION | | Authors: | Kandias, N.G, Chasapis, C.T, Bentrop, D, Episkopou, V, Spyroulias, G.A. | | Deposit date: | 2009-05-13 | | Release date: | 2010-05-19 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | NMR-based insights into the conformational and interaction properties of Arkadia RING-H2 E3 Ub ligase.
Proteins, 80, 2012
|
|
7P27
 
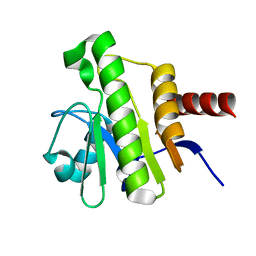 | | NMR solution structure of Chikungunya virus macro domain | | Descriptor: | Polyprotein P1234 | | Authors: | Lykouras, M.V, Tsika, A.C, Papageorgiou, N, Canard, B, Coutard, B, Bentrop, D, Spyroulias, G.A. | | Deposit date: | 2021-07-04 | | Release date: | 2022-07-13 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Binding Adaptation of GS-441524 Diversifies Macro Domains and Downregulates SARS-CoV-2 de-MARylation Capacity.
J.Mol.Biol., 434, 2022
|
|
7P2O
 
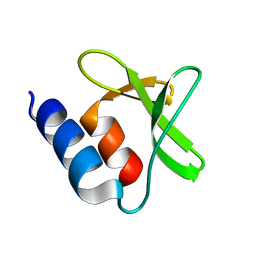 | |
