8T0J
 
 | | Salmonella Typhimurium ArnD | | 分子名称: | Probable 4-deoxy-4-formamido-L-arabinose-phosphoundecaprenol deformylase ArnD | | 著者 | Sousa, M.C, Munoz-Escudero, D, Lee, M. | | 登録日 | 2023-06-01 | | 公開日 | 2023-10-18 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.59 Å) | | 主引用文献 | Structure and Function of ArnD. A Deformylase Essential for Lipid A Modification with 4-Amino-4-deoxy-l-arabinose and Polymyxin Resistance.
Biochemistry, 62, 2023
|
|
1BUP
 
 | | T13S MUTANT OF BOVINE 70 KILODALTON HEAT SHOCK PROTEIN | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, MAGNESIUM ION, ... | | 著者 | Sousa, M.C, Mckay, D.B. | | 登録日 | 1998-09-03 | | 公開日 | 1998-09-09 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | The hydroxyl of threonine 13 of the bovine 70-kDa heat shock cognate protein is essential for transducing the ATP-induced conformational change.
Biochemistry, 37, 1998
|
|
8FTN
 
 | | E. coli ArnA dehydrogenase domain mutant - N492A | | 分子名称: | Bifunctional UDP-4-amino-4-deoxy-L-arabinose formyltransferase/UDP-glucuronic acid oxidase ArnA, SULFATE ION | | 著者 | Sousa, M.C, Mitchell, M.E. | | 登録日 | 2023-01-12 | | 公開日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Targeting the Conformational Change in ArnA Dehydrogenase for Selective Inhibition of Polymyxin Resistance.
Biochemistry, 62, 2023
|
|
8GJH
 
 | | Salmonella ArnA | | 分子名称: | Bifunctional polymyxin resistance protein ArnA, URIDINE-5'-DIPHOSPHATE-GLUCURONIC ACID | | 著者 | Sousa, M.C, Mitchell, M.E, Gatzeva-Topalova, P.Z. | | 登録日 | 2023-03-15 | | 公開日 | 2023-08-02 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (3.6 Å) | | 主引用文献 | Targeting the Conformational Change in ArnA Dehydrogenase for Selective Inhibition of Polymyxin Resistance.
Biochemistry, 62, 2023
|
|
1KYI
 
 | | HslUV (H. influenzae)-NLVS Vinyl Sulfone Inhibitor Complex | | 分子名称: | 4-IODO-3-NITROPHENYL ACETYL-LEUCINYL-LEUCINYL-LEUCINYL-VINYLSULFONE, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent hsl protease ATP-binding subunit hslU, ... | | 著者 | Sousa, M.C, Kessler, B.M, Overkleeft, H.S, McKay, D.B. | | 登録日 | 2002-02-04 | | 公開日 | 2002-05-15 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Crystal Structure of HslUV Complexed with a Vinyl Sulfone Inhibitor:
Corroboration of a Proposed Mechanism of Allosteric Activation of
HslV by HslU
J.Mol.Biol., 318, 2002
|
|
4LDR
 
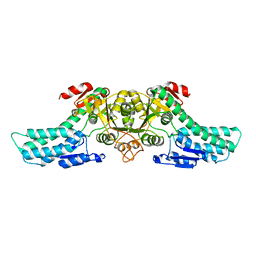 | |
4LDQ
 
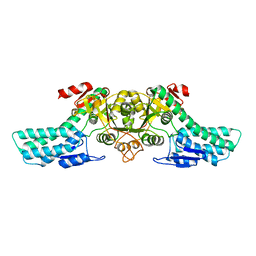 | |
2BUP
 
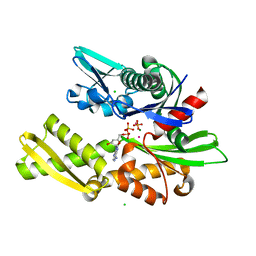 | | T13G Mutant of the ATPASE fragment of Bovine HSC70 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, ... | | 著者 | Sousa, M.C, Mckay, D.B. | | 登録日 | 1998-09-08 | | 公開日 | 1998-09-16 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | The hydroxyl of threonine 13 of the bovine 70-kDa heat shock cognate protein is essential for transducing the ATP-induced conformational change.
Biochemistry, 37, 1998
|
|
1G3I
 
 | | CRYSTAL STRUCTURE OF THE HSLUV PROTEASE-CHAPERONE COMPLEX | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, ATP-DEPENDENT HSLU PROTEASE ATP-BINDING SUBUNIT HSLU, ATP-DEPENDENT PROTEASE HSLV | | 著者 | Sousa, M.C, Trame, C.B, Tsuruta, H, Wilbanks, S.M, Reddy, V.S, McKay, D.B. | | 登録日 | 2000-10-24 | | 公開日 | 2000-11-22 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (3.41 Å) | | 主引用文献 | Crystal and solution structures of an HslUV protease-chaperone complex.
Cell(Cambridge,Mass.), 103, 2000
|
|
1G3K
 
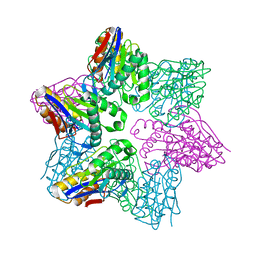 | |
1JJW
 
 | |
1JMV
 
 | |
4OCA
 
 | | Cryatal structure of ArnB K188A complexted with PLP and UDP-Ara4N | | 分子名称: | (2R,3R,4S,5S)-3,4-dihydroxy-5-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)amino]tetrahydro-2H-pyr an-2-yl [(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate, UDP-4-amino-4-deoxy-L-arabinose--oxoglutarate aminotransferase | | 著者 | Sousa, M.C, Lee, M. | | 登録日 | 2014-01-08 | | 公開日 | 2014-03-12 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural Basis for Substrate Specificity in ArnB. A Key Enzyme in the Polymyxin Resistance Pathway of Gram-Negative Bacteria.
Biochemistry, 53, 2014
|
|
4Q3J
 
 | |
1U2M
 
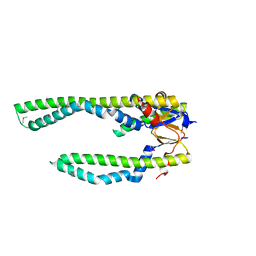 | | Crystal Structure of Skp | | 分子名称: | Histone-like protein HLP-1 | | 著者 | Walton, T.A, Sousa, M.C. | | 登録日 | 2004-07-19 | | 公開日 | 2004-08-24 | | 最終更新日 | 2017-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal Structure of Skp, a Prefoldin-like Chaperone that Protects Soluble and Membrane Proteins from Aggregation
Mol.Cell, 15, 2004
|
|
1U9J
 
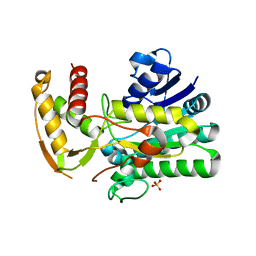 | |
1YRW
 
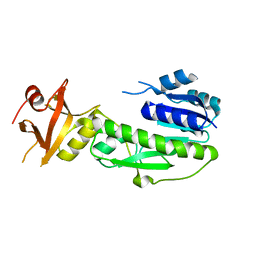 | |
3EFC
 
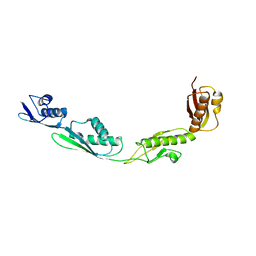 | |
4HDJ
 
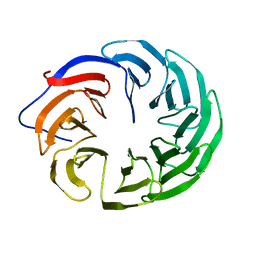 | |
3OG5
 
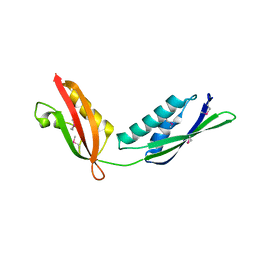 | | Crystal Structure of BamA POTRA45 tandem | | 分子名称: | Outer membrane protein assembly complex, YaeT protein | | 著者 | Gatzeva-Topalova, P.Z, Warner, L.R, Pardi, A, Sousa, M.C. | | 登録日 | 2010-08-16 | | 公開日 | 2010-11-17 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.69 Å) | | 主引用文献 | Structure and Flexibility of the Complete Periplasmic Domain of BamA: The Protein Insertion Machine of the Outer Membrane
Structure, 18, 2010
|
|
3QKY
 
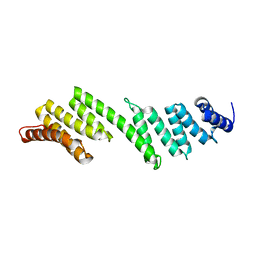 | | Crystal structure of Rhodothermus marinus BamD | | 分子名称: | Outer membrane assembly lipoprotein YfiO | | 著者 | Sandoval, C.M, Baker, S.L, Jansen, K, Metzner, S.I, Sousa, M.C. | | 登録日 | 2011-02-02 | | 公開日 | 2011-04-20 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Crystal structure of BamD: an essential component of the beta-Barrel assembly machinery of gram-negative bacteria.
J.Mol.Biol., 409, 2011
|
|
4PK1
 
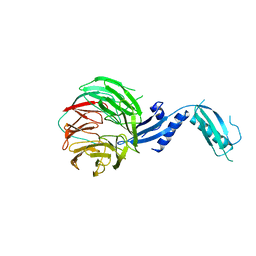 | | Structure of BamB fused to a BamA POTRA domain fragment | | 分子名称: | Chimera protein of Outer membrane protein assembly factors BamA and BamB | | 著者 | Jansen, K.B, Sousa, M.C. | | 登録日 | 2014-05-13 | | 公開日 | 2014-12-10 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Crystal Structure of BamB Bound to a Periplasmic Domain Fragment of BamA, the Central Component of the beta-Barrel Assembly Machine.
J.Biol.Chem., 290, 2015
|
|
1Z73
 
 | | Crystal Structure of E. coli ArnA dehydrogenase (decarboxylase) domain, S433A mutant | | 分子名称: | GLYCEROL, SULFATE ION, protein ArnA | | 著者 | Gatzeva-Topalova, P.Z, May, A.P, Sousa, M.C. | | 登録日 | 2005-03-24 | | 公開日 | 2005-06-07 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structure and Mechanism of ArnA: Conformational Change Implies Ordered Dehydrogenase Mechanism in Key Enzyme for Polymyxin Resistance
Structure, 13, 2005
|
|
1Z7B
 
 | |
1Z7E
 
 | | Crystal structure of full length ArnA | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, URIDINE-5'-DIPHOSPHATE-GLUCURONIC ACID, protein ArnA | | 著者 | Gatzeva-Topalova, P.Z, May, A.P, Sousa, M.C. | | 登録日 | 2005-03-24 | | 公開日 | 2005-06-07 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structure and Mechanism of ArnA: Conformational Change Implies Ordered Dehydrogenase Mechanism in Key Enzyme for Polymyxin Resistance
Structure, 13, 2005
|
|
