7QDH
 
 | | SARS-CoV-2 S protein S:D614G mutant 1-up | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin | | Authors: | Ginex, T, Marco-Marin, C, Wieczor, M, Mata, C.P, Krieger, J, Lopez-Redondo, M.L, Frances-Gomez, C, Ruiz-Rodriguez, P, Melero, R, Sanchez-Sorzano, C.O, Martinez, M, Gougeard, N, Forcada-Nadal, A, Zamora-Caballero, S, Gozalbo-Rovira, R, Sanz-Frasquet, C, Bravo, J, Rubio, V, Marina, A, Geller, R, Comas, I, Gil, C, Coscolla, M, Orozco, M, LLacer, J.L, Carazo, J.M. | | Deposit date: | 2021-11-27 | | Release date: | 2022-05-25 | | Last modified: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | The structural role of SARS-CoV-2 genetic background in the emergence and success of spike mutations: The case of the spike A222V mutation.
Plos Pathog., 18, 2022
|
|
7QDG
 
 | | SARS-CoV-2 S protein S:A222V + S:D614G mutant 1-up | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ginex, T, Marco-Marin, C, Wieczor, M, Mata, C.P, Krieger, J, Lopez-Redondo, M.L, Frances-Gomez, C, Ruiz-Rodriguez, P, Melero, R, Sanchez-Sorzano, C.O, Martinez, M, Gougeard, N, Forcada-Nadal, A, Zamora-Caballero, S, Gozalbo-Rovira, R, Sanz-Frasquet, C, Bravo, J, Rubio, V, Marina, A, Geller, R, Comas, I, Gil, C, Coscolla, M, Orozco, M, LLacer, J.L, Carazo, J.M. | | Deposit date: | 2021-11-27 | | Release date: | 2022-05-25 | | Last modified: | 2022-08-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The structural role of SARS-CoV-2 genetic background in the emergence and success of spike mutations: The case of the spike A222V mutation.
Plos Pathog., 18, 2022
|
|
4BLF
 
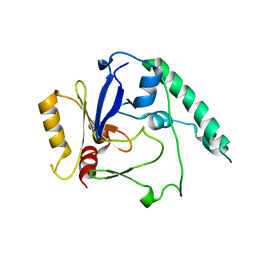 | | Variable internal flexibility characterizes the helical capsid formed by Agrobacterium VirE2 protein on single-stranded DNA. | | Descriptor: | SINGLE-STRAND DNA-BINDING PROTEIN | | Authors: | Bharat, T.A.M, Zbaida, D, Eisenstein, M, Frankenstein, Z, Mehlman, T, Weiner, L, Sorzano, C.O.S, Barak, Y, Albeck, S, Briggs, J.A.G, Wolf, S.G, Elbaum, M. | | Deposit date: | 2013-05-02 | | Release date: | 2013-06-26 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | Variable Internal Flexibility Characterizes the Helical Capsid Formed by Agrobacterium Vire2 Protein on Single-Stranded DNA.
Structure, 21, 2013
|
|
7QCO
 
 | | The structure of Photosystem I tetramer from Chroococcidiopsis TS-821, a thermophilic, unicellular, non-heterocyst-forming cyanobacterium | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, BETA-CAROTENE, CHLOROPHYLL A, ... | | Authors: | Semchonok, D.A, Mondal, J, Cooper, J.C, Schlum, K, Li, M, Amin, M, Sorzano, C.O.S, Ramirez-Aportela, E, Kastritis, P.L, Boekema, E.J, Guskov, A, Bruce, B.D. | | Deposit date: | 2021-11-24 | | Release date: | 2022-04-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structure of a tetrameric photosystem I from Chroococcidiopsis TS-821, a thermophilic, unicellular, non-heterocyst-forming cyanobacterium.
Plant Commun., 3, 2022
|
|
6I7M
 
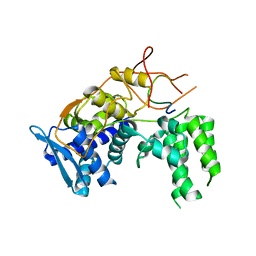 | | Influenza A nucleoprotein docked into 3D helical structure of the wild type ribonucleoprotein complex obtained using cryoEM. Conformation 4. | | Descriptor: | Nucleoprotein | | Authors: | Coloma, R, Arranz, R, de la Rosa-Trevin, J.M, Sorzano, C.O.S, Carlero, D, Ortin, J, Martin-Benito, J. | | Deposit date: | 2018-11-16 | | Release date: | 2020-02-12 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Structural insights into influenza A virus ribonucleoproteins reveal a processive helical track as transcription mechanism.
Nat Microbiol, 5, 2020
|
|
6I7B
 
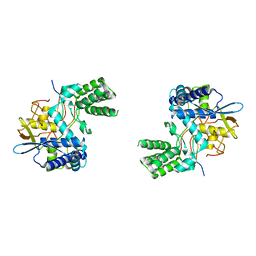 | | Influenza A nucleoprotein docked into 3D helical structure of the wild type ribonucleoprotein complex obtained using cryoEM. Conformation 3. | | Descriptor: | Nucleoprotein | | Authors: | Coloma, R, Arranz, R, de la Rosa-Trevin, J.M, Sorzano, C.O.S, Carlero, D, Ortin, J, Martin-Benito, J. | | Deposit date: | 2018-11-16 | | Release date: | 2020-02-19 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Structural insights into influenza A virus ribonucleoproteins reveal a processive helical track as transcription mechanism.
Nat Microbiol, 5, 2020
|
|
6H9G
 
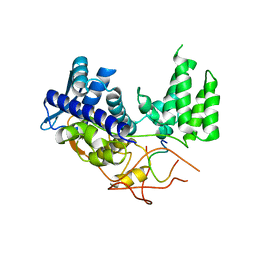 | | Influenza A nucleoprotein docked into 3D helical structure of the wild type ribonucleoprotein complex obtained using cryoEM. Conformation 1. | | Descriptor: | Nucleoprotein, Polypeptide loop | | Authors: | Coloma, R, Arranz, R, de la Rosa-Trevin, J.M, Sorzano, C.O.S, Munier, S, Carlero, D, Naffakh, N, Ortin, J, Martin-Benito, J. | | Deposit date: | 2018-08-03 | | Release date: | 2020-02-12 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (11 Å) | | Cite: | Structural insights into influenza A virus ribonucleoproteins reveal a processive helical track as transcription mechanism.
Nat Microbiol, 5, 2020
|
|
6I85
 
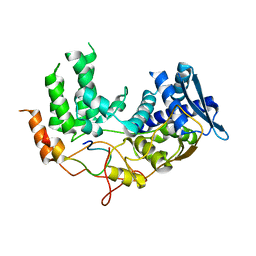 | | Influenza A nucleoprotein docked into the 3D helical structure of the wild type ribonucleoprotein complex obtained using cryoEM. Conformation 5. | | Descriptor: | Influenza A nucleoprotein, Nucleoprotein | | Authors: | Coloma, R, Arranz, R, de la Rosa-Trevin, J.M, Sorzano, C.O.S, Carlero, D, Ortin, J, Martin-Benito, J. | | Deposit date: | 2018-11-19 | | Release date: | 2020-01-29 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (24 Å) | | Cite: | Structural insights into influenza A virus ribonucleoproteins reveal a processive helical track as transcription mechanism.
Nat Microbiol, 5, 2020
|
|
6I54
 
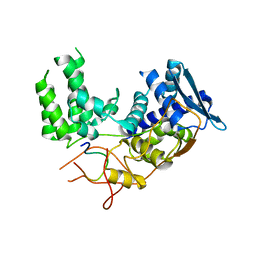 | | Influenza A nucleoprotein docked into 3D helical structure of the wild type ribonucleoprotein complex obtained using cryoEM. Conformation 2. | | Descriptor: | Influenza virus nucleoprotein, Nucleoprotein | | Authors: | Coloma, R, Arranz, R, de la Rosa-Trevin, J.M, Sorzano, C.O.S, Carlero, D, Ortin, J, Martin-Benito, J. | | Deposit date: | 2018-11-12 | | Release date: | 2019-11-13 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Structural insights into influenza A virus ribonucleoproteins reveal a processive helical track as transcription mechanism.
Nat Microbiol, 5, 2020
|
|
