7BSD
 
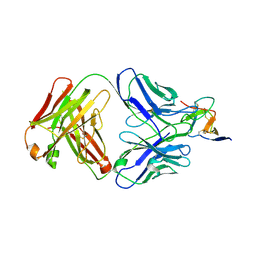 | |
7BSC
 
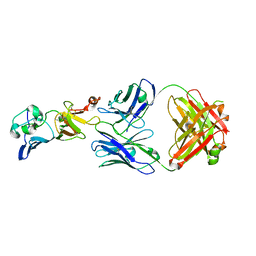 | | Complex structure of 1G5.3 Fab bound to DENV2 NS1c | | Descriptor: | 1G5.3 Fab Heavy Chain, 1G5.3 Fab Light Chain, Non-structural protein 1 | | Authors: | Song, H, Qi, J, Gao, F.G. | | Deposit date: | 2020-03-30 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | A broadly protective antibody that targets the flavivirus NS1 protein.
Science, 371, 2021
|
|
7ZB0
 
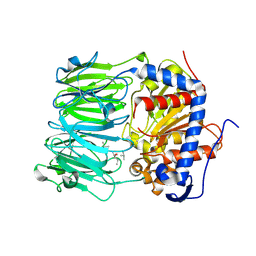 | | macrocyclase OphP with 15mer | | Descriptor: | 1,2-ETHANEDIOL, 15mer, BICARBONATE ION, ... | | Authors: | Song, H, Naismith, J.H. | | Deposit date: | 2022-03-23 | | Release date: | 2022-07-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Molecular basis for the enzymatic macrocyclization of multiply backbone N-methylated peptides
Biorxiv, 2022
|
|
7ZB1
 
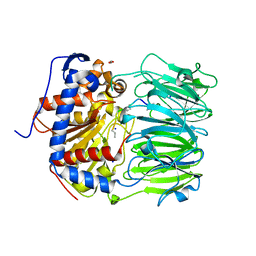 | | S580A with 18mer | | Descriptor: | 1,2-ETHANEDIOL, 18mer, BICARBONATE ION, ... | | Authors: | Song, H, Naismith, J.H. | | Deposit date: | 2022-03-23 | | Release date: | 2022-07-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular basis for the enzymatic macrocyclization of multiply backbone N-methylated peptides
Biorxiv, 2022
|
|
7ZAZ
 
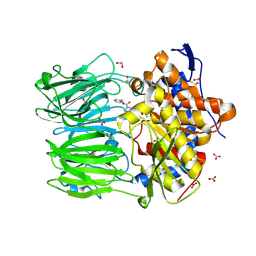 | | macrocyclase OphP with ZPP | | Descriptor: | 1,2-ETHANEDIOL, BICARBONATE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Song, H, Naismith, J.H. | | Deposit date: | 2022-03-23 | | Release date: | 2022-07-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular basis for the enzymatic macrocyclization of multiply backbone N-methylated peptides
Biorxiv, 2022
|
|
1KCT
 
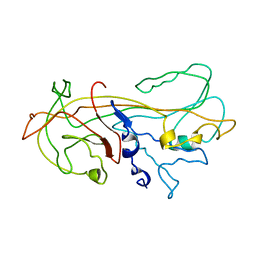 | | ALPHA1-ANTITRYPSIN | | Descriptor: | ALPHA1-ANTITRYPSIN | | Authors: | Song, H.K, Suh, S.W. | | Deposit date: | 1996-08-06 | | Release date: | 1997-01-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.46 Å) | | Cite: | Crystal structure of an uncleaved alpha 1-antitrypsin reveals the conformation of its inhibitory reactive loop.
FEBS Lett., 377, 1995
|
|
4EML
 
 | |
7ZB2
 
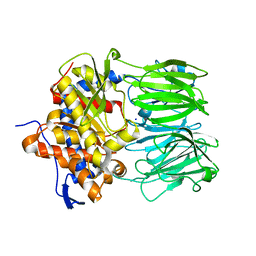 | | apo macrocyclase OphP | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Song, H, Naismith, J.H. | | Deposit date: | 2022-03-23 | | Release date: | 2022-07-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Molecular basis for the enzymatic macrocyclization of multiply backbone N-methylated peptides
Biorxiv, 2022
|
|
1OX9
 
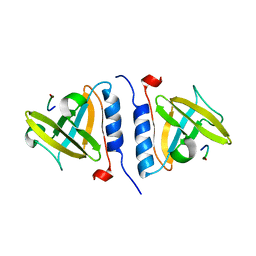 | | Crystal structure of SspB-ssrA complex | | Descriptor: | Stringent starvation protein B, ssrA | | Authors: | Song, H.K, Eck, M.J. | | Deposit date: | 2003-04-01 | | Release date: | 2003-08-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of degradation signal recognition by SspB, a specificity-enhancing factor for the ClpXP proteolytic machine
Mol.Cell, 12, 2003
|
|
6GEW
 
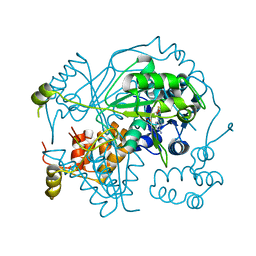 | | OphA Y63F-sinefungin complex | | Descriptor: | OphA, S-ADENOSYL-L-HOMOCYSTEINE, SINEFUNGIN | | Authors: | Song, H, Naismith, J.H. | | Deposit date: | 2018-04-27 | | Release date: | 2018-09-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A molecular mechanism for the enzymatic methylation of nitrogen atoms within peptide bonds.
Sci Adv, 4, 2018
|
|
1GMV
 
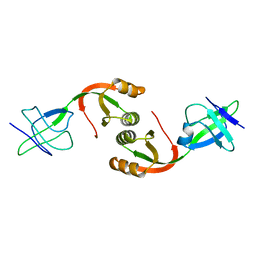 | | Structure of UreE | | Descriptor: | UREE | | Authors: | Song, H.K, Mulrooney, S.B, Huber, R, Hausinger, R. | | Deposit date: | 2001-09-24 | | Release date: | 2001-11-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Klebsiella Aerogenes Uree, a Nickel-Binding Metallochaperone for Urease Activation.
J.Biol.Chem., 276, 2001
|
|
1M4Y
 
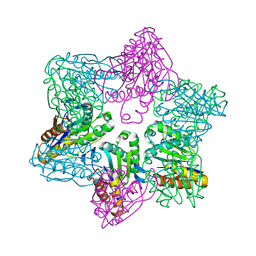 | | Crystal structure of HslV from Thermotoga maritima | | Descriptor: | ATP-dependent protease hslV, SODIUM ION | | Authors: | Song, H.K, Ramachandran, R, Bochtler, M.B, Hartmann, C, Azim, M.K, Huber, R. | | Deposit date: | 2002-07-05 | | Release date: | 2003-05-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Isolation and characterization of the prokaryotic proteasome homolog HslVU (ClpQY) from Thermotoga maritima and the crystal structure of HslV.
BIOPHYS.CHEM., 100, 2003
|
|
4I4Z
 
 | | Synechocystis sp. PCC 6803 1,4-dihydroxy-2-naphthoyl-coenzyme A synthase (MenB) in complex with salicylyl-CoA | | Descriptor: | BICARBONATE ION, MALONATE ION, Naphthoate synthase, ... | | Authors: | Song, H.G, Sun, Y.R, Li, J, Li, Y, Jiang, M, Zhou, J.H, Guo, Z.H. | | Deposit date: | 2012-11-28 | | Release date: | 2013-05-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of the induced-fit mechanism of 1,4-dihydroxy-2-naphthoyl coenzyme a synthase from the crotonase fold superfamily
Plos One, 8, 2013
|
|
1VJS
 
 | |
2WSK
 
 | | Crystal structure of Glycogen Debranching Enzyme GlgX from Escherichia coli K-12 | | Descriptor: | GLYCOGEN DEBRANCHING ENZYME, SULFATE ION | | Authors: | Song, H.-N, Park, J.-T, Jung, T.-Y, Park, K.-H, Woo, E.-J. | | Deposit date: | 2009-09-08 | | Release date: | 2010-09-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural Rationale for the Short Branched Substrate Specificity of the Glycogen Debranching Enzyme Glgx.
Proteins, 78, 2010
|
|
4I52
 
 | | scMenB im complex with 1-hydroxy-2-naphthoyl-CoA | | Descriptor: | 1-hydroxy-2-naphthoyl-CoA, CHLORIDE ION, Naphthoate synthase | | Authors: | Song, H.G, Sun, Y.R, Li, J, Li, Y, Jiang, M, Zhou, J.H, Guo, Z.H. | | Deposit date: | 2012-11-28 | | Release date: | 2013-05-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis of the induced-fit mechanism of 1,4-dihydroxy-2-naphthoyl coenzyme a synthase from the crotonase fold superfamily
Plos One, 8, 2013
|
|
1T91
 
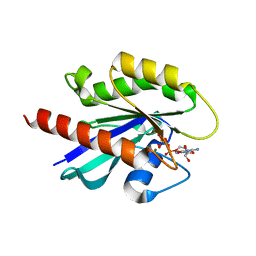 | | crystal structure of human small GTPase Rab7(GTP) | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Ras-related protein Rab-7 | | Authors: | Song, H, Hong, W, Wu, M, Wang, T. | | Deposit date: | 2004-05-14 | | Release date: | 2005-05-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for recruitment of RILP by small GTPase Rab7.
Embo J., 24, 2005
|
|
2X6R
 
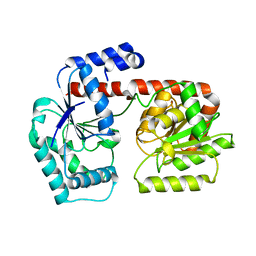 | | Crystal structure of trehalose synthase TreT from P.horikoshi produced by soaking in trehalose | | Descriptor: | TREHALOSE-SYNTHASE TRET | | Authors: | Song, H.-N, Jung, T.-Y, Yoon, S.-M, Lim, M.-Y, Lee, S.-B, Woo, E.-J. | | Deposit date: | 2010-02-19 | | Release date: | 2010-10-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Insights on the New Mechanism of Trehalose Synthesis by Trehalose Synthase Tret from Pyrococcus Horikoshii.
J.Mol.Biol., 404, 2010
|
|
2XA9
 
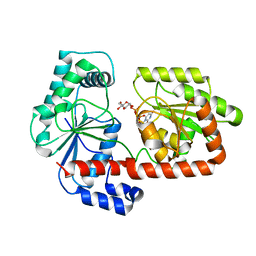 | | Crystal structure of trehalose synthase TreT mutant E326A from P. horikoshii in complex with UDPG | | Descriptor: | TREHALOSE-SYNTHASE TRET, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Song, H.-N, Jung, T.-Y, Yoon, S.-M, Lee, S.-B, Lim, M.-Y, Woo, E.-J. | | Deposit date: | 2010-03-30 | | Release date: | 2011-03-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Insights on the New Mechanism of Trehalose Synthesis by Trehalose Synthase Tret from Pyrococcus Horikoshii.
J.Mol.Biol., 404, 2010
|
|
2X6Q
 
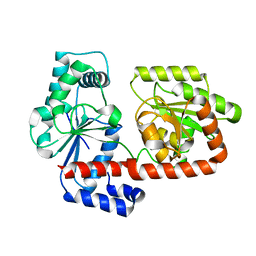 | | Crystal structure of trehalose synthase TreT from P.horikoshi | | Descriptor: | TREHALOSE-SYNTHASE TRET | | Authors: | Song, H.-N, Jung, T.-Y, Yoon, S.-M, Lim, M.-Y, Lee, S.-B, Woo, E.-J. | | Deposit date: | 2010-02-19 | | Release date: | 2010-10-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Insights on the New Mechanism of Trehalose Synthesis by Trehalose Synthase Tret from Pyrococcus Horikoshii.
J.Mol.Biol., 404, 2010
|
|
2XA1
 
 | | Crystal structure of trehalose synthase TreT from P.horikoshii (Seleno derivative) | | Descriptor: | TREHALOSE-SYNTHASE TRET | | Authors: | Song, H.-N, Jung, T.-Y, Yoon, S.-M, Lee, S.-B, Lim, M.-Y, Woo, E.-J. | | Deposit date: | 2010-03-26 | | Release date: | 2010-10-13 | | Last modified: | 2012-06-27 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Structural Insights on the New Mechanism of Trehalose Synthesis by Trehalose Synthase Tret from Pyrococcus Horikoshii.
J.Mol.Biol., 404, 2010
|
|
2XA2
 
 | | Crystal structure of trehalose synthase TreT mutant E326A from P. horikoshii in complex with UDPG | | Descriptor: | TREHALOSE-SYNTHASE TRET, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Song, H.-N, Jung, T.-Y, Yoon, S.-M, Lee, S.-B, Lim, M.-Y, Woo, E.-J. | | Deposit date: | 2010-03-26 | | Release date: | 2011-03-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Insights on the New Mechanism of Trehalose Synthesis by Trehalose Synthase Tret from Pyrococcus Horikoshii.
J.Mol.Biol., 404, 2010
|
|
2XMP
 
 | | Crystal structure of trehalose synthase TreT mutant E326A from P. horishiki in complex with UDP | | Descriptor: | TREHALOSE-SYNTHASE TRET, URIDINE-5'-DIPHOSPHATE | | Authors: | Song, H.-N, Jung, T.-Y, Yoon, S.-M, Lee, S.-B, Lim, M.-Y, Woo, E.-J. | | Deposit date: | 2010-07-29 | | Release date: | 2010-10-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Insights on the New Mechanism of Trehalose Synthesis by Trehalose Synthase Tret from Pyrococcus Horikoshii.
J.Mol.Biol., 404, 2010
|
|
2DS5
 
 | | Structure of the ZBD in the orthorhomibic crystal from | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit clpX, CALCIUM ION, TETRAETHYLENE GLYCOL, ... | | Authors: | Song, H.K, Park, E.Y, Lee, B.G, Hong, S.B. | | Deposit date: | 2006-06-22 | | Release date: | 2007-02-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Basis of SspB-tail Recognition by the Zinc Binding Domain of ClpX.
J.Mol.Biol., 367, 2007
|
|
4AEF
 
 | | THE CRYSTAL STRUCTURE OF THERMOSTABLE AMYLASE FROM THE PYROCOCCUS | | Descriptor: | NEOPULLULANASE (ALPHA-AMYLASE II) | | Authors: | Song, H.-N, Jung, T.-Y, Yoon, S.-M, Yang, S.-J, Park, K.-H, Woo, E.-J. | | Deposit date: | 2012-01-10 | | Release date: | 2012-10-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | A Novel Domain Arrangement in a Monomeric Cyclodextrin-Hydrolyzing Enzyme from the Hyperthermophile Pyrococcus Furiosus.
Biochim.Biophys.Acta, 1834, 2013
|
|
