5ROB
 
 | | PanDDA analysis group deposition of ground-state model of SARS-CoV-2 helicase | | 分子名称: | Helicase, PHOSPHATE ION, ZINC ION | | 著者 | Newman, J.A, Yosaatmadja, Y, Douangamath, A, Aimon, A, Powell, A.J, Dias, A, Fearon, D, Dunnett, L, Brandao-Neto, J, Krojer, T, Skyner, R, Gorrie-Stone, T, Thompson, W, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | 登録日 | 2020-09-22 | | 公開日 | 2021-03-10 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.87 Å) | | 主引用文献 | PanDDA analysis group deposition of ground-state model
To Be Published
|
|
5S73
 
 | | PanDDA analysis group deposition of ground-state model of SARS-CoV-2 Nsp3 macrodomain | | 分子名称: | Non-structural protein 3 | | 著者 | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | 登録日 | 2020-11-23 | | 公開日 | 2021-01-13 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.06 Å) | | 主引用文献 | PanDDA analysis group deposition of ground-state model of SARS-CoV-2 Nsp3 macrodomain
To Be Published
|
|
5S74
 
 | | PanDDA analysis group deposition of ground-state model of SARS-CoV-2 Nsp3 macrodomain | | 分子名称: | 1,2-ETHANEDIOL, Non-structural protein 3 | | 著者 | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | 登録日 | 2020-11-23 | | 公開日 | 2021-01-13 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (0.96 Å) | | 主引用文献 | PanDDA analysis group deposition of ground-state model of SARS-CoV-2 Nsp3 macrodomain
To Be Published
|
|
5X13
 
 | | Crystal structure of Bacillus subtilis PadR in complex with p-coumaric acid | | 分子名称: | 4'-HYDROXYCINNAMIC ACID, GLYCEROL, Transcriptional regulator | | 著者 | Park, S.C, Kwak, Y.M, Song, W.S, Hong, M, Yoon, S.I. | | 登録日 | 2017-01-24 | | 公開日 | 2017-11-22 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural basis of effector and operator recognition by the phenolic acid-responsive transcriptional regulator PadR
Nucleic Acids Res., 45, 2017
|
|
4NEA
 
 | | 1.90 Angstrom resolution crystal structure of betaine aldehyde dehydrogenase (betB) from Staphylococcus aureus in complex with NAD+ and BME-free Cys289 | | 分子名称: | Betaine aldehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, POTASSIUM ION | | 著者 | Halavaty, A.S, Minasov, G, Winsor, J, Dubrovska, I, Shuvalova, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2013-10-28 | | 公開日 | 2013-11-06 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural and functional analysis of betaine aldehyde dehydrogenase from Staphylococcus aureus.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4NOG
 
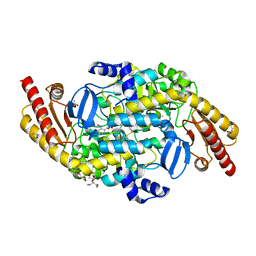 | | Crystal structure of a putative ornithine aminotransferase from Toxoplasma gondii ME49 in complex with pyrodoxal-5'-phosphate | | 分子名称: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, BETA-MERCAPTOETHANOL, ... | | 著者 | Filippova, E.V, Halavaty, A, Ruan, J, Shuvalova, L, Flores, K, Dubrovska, I, Ngo, H, Shanmugam, D, Roos, D, Anderson, W.F, Center for Structural Genomics of Infectious Diseases, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2013-11-19 | | 公開日 | 2013-12-04 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | CSGID Solves Structures and Identifies Phenotypes for Five Enzymes in Toxoplasma gondii .
Front Cell Infect Microbiol, 8, 2018
|
|
5I0D
 
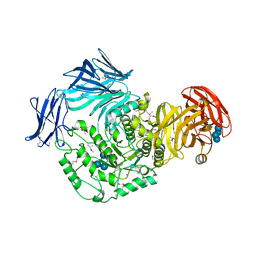 | | Cycloalternan-forming enzyme from Listeria monocytogenes in complex with cycloalternan | | 分子名称: | CALCIUM ION, CHLORIDE ION, Cyclic alpha-D-glucopyranose-(1-3)-alpha-D-glucopyranose-(1-6)-alpha-D-glucopyranose-(1-3)-alpha-D-glucopyranose, ... | | 著者 | Light, S.H, Minasov, G, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2016-02-03 | | 公開日 | 2016-12-14 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.77 Å) | | 主引用文献 | Transferase Versus Hydrolase: The Role of Conformational Flexibility in Reaction Specificity.
Structure, 25, 2017
|
|
5E31
 
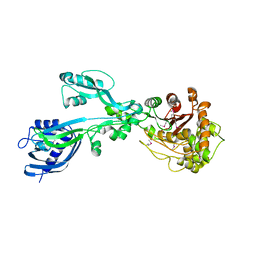 | | 2.3 Angstrom Crystal Structure of the Monomeric Form of Penicillin Binding Protein 2 Prime from Enterococcus faecium. | | 分子名称: | Penicillin binding protein 2 prime | | 著者 | Minasov, G, Wawrzak, Z, Shuvalova, L, Dubrovska, I, Flores, K, Filippova, E, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2015-10-01 | | 公開日 | 2015-10-14 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | 2.3 Angstrom Crystal Structure of the Monomeric Form of Penicillin Binding Protein 2 Prime from Enterococcus faecium.
To Be Published
|
|
4OHN
 
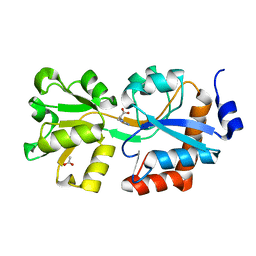 | | Crystal structure of an ABC uptake transporter substrate binding protein from Streptococcus pneumoniae with Bound Histidine | | 分子名称: | ABC transporter substrate-binding protein, ACETATE ION, HISTIDINE | | 著者 | Brunzelle, J.S, Wawrzak, W, Yim, Y, Kudritska, M, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2014-01-17 | | 公開日 | 2015-01-21 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.37 Å) | | 主引用文献 | Crystal structure of an ABC uptake transporter substrate binding protein from Streptococcus pneumoniae with Bound Histidine
To be Published
|
|
7UQC
 
 | |
5XLJ
 
 | | Crystal structure of the flagellar cap protein flid D2-D3 domains from serratia marcescens in Space group P432 | | 分子名称: | CHLORIDE ION, Flagellar hook-associated protein 2, SODIUM ION | | 著者 | Cho, S.Y, Song, W.S, Hong, H.J, Yoon, S.I. | | 登録日 | 2017-05-10 | | 公開日 | 2017-06-14 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Tetrameric structure of the flagellar cap protein FliD from Serratia marcescens.
Biochem. Biophys. Res. Commun., 489, 2017
|
|
5XLR
 
 | |
5XLK
 
 | | Crystal structure of the flagellar cap protein FliD D2-D3 domains from Serratia marcescens in Space group I422 | | 分子名称: | Flagellar hook-associated protein 2, ZINC ION | | 著者 | Cho, S.Y, Song, W.S, Hong, H.J, Yoon, S.I. | | 登録日 | 2017-05-10 | | 公開日 | 2017-06-14 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.05 Å) | | 主引用文献 | Tetrameric structure of the flagellar cap protein FliD from Serratia marcescens.
Biochem. Biophys. Res. Commun., 489, 2017
|
|
5F5X
 
 | |
5F2H
 
 | | 2.75 Angstrom resolution crystal structure of uncharacterized protein from Bacillus cereus ATCC 10987 | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Uncharacterized protein | | 著者 | Halavaty, A.S, Filippova, E.V, Wawrzak, Z, Minasov, G, Kiryukhina, O, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2015-12-01 | | 公開日 | 2015-12-16 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | 2.75 Angstrom resolution crystal structure of uncharacterized protein from Bacillus cereus ATCC 10987
To Be Published
|
|
5EV7
 
 | | The crystal structure of a functionally unknown conserved protein mutant from Bacillus anthracis str. Ames | | 分子名称: | Conserved domain protein | | 著者 | Tan, K, Zhou, M, Jedrzejczak, R, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2015-11-19 | | 公開日 | 2015-12-09 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.351 Å) | | 主引用文献 | The crystal structure of a functionally unknown conserved protein mutant from Bacillus anthracis str. Ames.
To Be Published
|
|
5EWQ
 
 | | The crystal structure of an amidase family protein from Bacillus anthracis str. Ames | | 分子名称: | ACETATE ION, Amidase | | 著者 | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2015-11-20 | | 公開日 | 2015-12-09 | | 最終更新日 | 2019-12-04 | | 実験手法 | X-RAY DIFFRACTION (2.57 Å) | | 主引用文献 | The crystal structure of an amidase family protein from Bacillus anthracis str. Ames
To Be Published
|
|
6MUQ
 
 | | 1.67 Angstrom Resolution Crystal Structure of Murein-DD-endopeptidase from Yersinia enterocolitica. | | 分子名称: | ACETATE ION, Murein-DD-endopeptidase, SULFATE ION | | 著者 | Minasov, G, Shuvalova, L, Kiryukhina, O, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2018-10-23 | | 公開日 | 2018-10-31 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.67 Å) | | 主引用文献 | 1.67 Angstrom Resolution Crystal Structure of Murein-DD-endopeptidase from Yersinia enterocolitica.
To Be Published
|
|
5F7S
 
 | |
227D
 
 | | A CRYSTALLOGRAPHIC AND SPECTROSCOPIC STUDY OF THE COMPLEX BETWEEN D(CGCGAATTCGCG)2 AND 2,5-BIS(4-GUANYLPHENYL)FURAN, AN ANALOGUE OF BERENIL. STRUCTURAL ORIGINS OF ENHANCED DNA-BINDING AFFINITY | | 分子名称: | 2,5-BIS(4-GUANYLPHENYL)FURAN, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3') | | 著者 | Laughton, C.A, Tanious, F, Nunn, C.M, Boykin, D.W, Wilson, W.D, Neidle, S. | | 登録日 | 1995-08-08 | | 公開日 | 1995-11-11 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | A crystallographic and spectroscopic study of the complex between d(CGCGAATTCGCG)2 and 2,5-bis(4-guanylphenyl)furan, an analogue of berenil. Structural origins of enhanced DNA-binding affinity.
Biochemistry, 35, 1996
|
|
5YHH
 
 | | Crystal structure of YiiM from Geobacillus stearothermophilus | | 分子名称: | Uncharacterized conserved protein YiiM | | 著者 | Namgung, B, Kim, J.H, Song, W.S, Yoon, S.I. | | 登録日 | 2017-09-28 | | 公開日 | 2018-03-07 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of the hydroxylaminopurine resistance protein, YiiM, and its putative molybdenum cofactor-binding catalytic site.
Sci Rep, 8, 2018
|
|
5JPI
 
 | | 2.15 Angstrom Crystal Structure of S-adenosylhomocysteinase from Cryptosporidium parvum in Complex with D-Eritadenine and NAD | | 分子名称: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Adenosylhomocysteinase, ... | | 著者 | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Bishop, B, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2016-05-03 | | 公開日 | 2016-05-18 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | 2.15 Angstrom Crystal Structure of S-adenosylhomocysteinase from Cryptosporidium parvum in Complex with D-Eritadenine and NAD.
To Be Published
|
|
3IRH
 
 | | Structure of an Enterococcus Faecalis HD-domain protein complexed with dGTP and dATP | | 分子名称: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, CALCIUM ION, ... | | 著者 | Vorontsov, I.I, Minasov, G, Shuvalova, L, Brunzelle, J.S, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2009-08-24 | | 公開日 | 2009-09-22 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Characterization of the deoxynucleotide triphosphate triphosphohydrolase (dNTPase) activity of the EF1143 protein from Enterococcus faecalis and crystal structure of the activator-substrate complex.
J.Biol.Chem., 286, 2011
|
|
5DZS
 
 | | 1.5 Angstrom Crystal Structure of Shikimate Dehydrogenase 1 from Peptoclostridium difficile. | | 分子名称: | SULFATE ION, Shikimate dehydrogenase (NADP(+)) | | 著者 | Minasov, G, Wawrzak, Z, Shuvalova, L, Dubrovska, I, Flores, K, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2015-09-26 | | 公開日 | 2015-10-07 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | 1.5 Angstrom Crystal Structure of Shikimate Dehydrogenase 1 from Peptoclostridium difficile.
To Be Published
|
|
5YHI
 
 | | Crystal structure of YiiM from Escherichia coli | | 分子名称: | PHOSPHATE ION, Protein YiiM | | 著者 | Namgung, B, Kim, J.H, Song, W.S, Yoon, S.I. | | 登録日 | 2017-09-28 | | 公開日 | 2018-03-07 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | Crystal structure of the hydroxylaminopurine resistance protein, YiiM, and its putative molybdenum cofactor-binding catalytic site.
Sci Rep, 8, 2018
|
|
