1WIP
 
 | |
1WIO
 
 | |
2LRJ
 
 | |
1WIQ
 
 | |
1AXM
 
 | | HEPARIN-LINKED BIOLOGICALLY-ACTIVE DIMER OF FIBROBLAST GROWTH FACTOR | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid, 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, ACIDIC FIBROBLAST GROWTH FACTOR | | Authors: | DiGabriele, A.D, Lax, I, Chen, D.I, Svahn, C.M, Jaye, M, Schlessinger, J, Hendrickson, W.A. | | Deposit date: | 1997-10-16 | | Release date: | 1998-04-22 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of a heparin-linked biologically active dimer of fibroblast growth factor.
Nature, 393, 1998
|
|
1WP0
 
 | | Human SCO1 | | Descriptor: | SCO1 protein homolog | | Authors: | Williams, J.C, Sue, C, Banting, G.S, Yang, H, Glerum, D.M, Hendrickson, W.A, Schon, E.A. | | Deposit date: | 2004-08-27 | | Release date: | 2005-01-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Human SCO1: IMPLICATIONS FOR REDOX SIGNALING BY A MITOCHONDRIAL CYTOCHROME c OXIDASE "ASSEMBLY" PROTEIN
J.Biol.Chem., 280, 2005
|
|
2M1Z
 
 | |
1XA0
 
 | | Crystal Structure of MCSG Target APC35536 from Bacillus stearothermophilus | | Descriptor: | CHLORIDE ION, Putative NADPH Dependent oxidoreductases, SULFATE ION | | Authors: | Brunzelle, J.S, Sommerhalter, M, Minasov, G, Shuvalova, L, Collart, F.R, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-08-24 | | Release date: | 2004-10-05 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of MCSG Target APC35536 from Bacillus stearothermophilus
To be Published
|
|
2I0Z
 
 | | Crystal structure of a FAD binding protein from Bacillus cereus, a putative NAD(FAD)-utilizing dehydrogenases | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, NAD(FAD)-utilizing dehydrogenases | | Authors: | Minasov, G, Shuvalova, L, Vorontsov, I.I, Kiryukhina, O, Abdullah, J, Collart, F.R, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-08-11 | | Release date: | 2006-08-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal structure of a FAD binding protein from Bacillus cereus, a putative NAD(FAD)-utilizing dehydrogenases
To be Published
|
|
1X7F
 
 | | Crystal structure of an uncharacterized B. cereus protein | | Descriptor: | outer surface protein | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Collart, F.R, Anderson, W.F, MCSG, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-08-13 | | Release date: | 2004-08-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of an uncharacterized B. cereus protein
To be Published
|
|
2NSB
 
 | |
2NSA
 
 | |
2NSC
 
 | |
2OL5
 
 | | Crystal Structure of a protease synthase and sporulation negative regulatory protein PAI 2 from Bacillus stearothermophilus | | Descriptor: | PAI 2 protein | | Authors: | Brunzelle, J.S, Cuff, M.E, Minasov, G, Li, H, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-01-18 | | Release date: | 2007-02-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the novel PaiB transcriptional regulator from Geobacillus stearothermophilus.
Proteins, 79, 2011
|
|
1R5L
 
 | | Crystal Structure of Human Alpha-Tocopherol Transfer Protein Bound to its Ligand | | Descriptor: | (2R)-2,5,7,8-TETRAMETHYL-2-[(4R,8R)-4,8,12-TRIMETHYLTRIDECYL]CHROMAN-6-OL, PROTEIN (Alpha-tocopherol transfer protein) | | Authors: | Min, K.C, Kovall, R.A, Hendrickson, W.A. | | Deposit date: | 2003-10-10 | | Release date: | 2003-11-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of human alpha-tocopherol transfer protein bound to its ligand: Implications for ataxia with vitamin E deficiency
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
2OKQ
 
 | | Crystal structure of unknown conserved ybaA protein from Shigella flexneri | | Descriptor: | Hypothetical protein ybaA, SODIUM ION | | Authors: | Minasov, G, Vorontsov, I.I, Shuvalova, L, Brunzelle, J.S, Kiryukhina, O, Collart, F.R, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-01-17 | | Release date: | 2007-02-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of unknown conserved ybaA protein from Shigella flexneri
TO BE PUBLISHED
|
|
1BUN
 
 | | STRUCTURE OF BETA2-BUNGAROTOXIN: POTASSIUM CHANNEL BINDING BY KUNITZ MODULES AND TARGETED PHOSPHOLIPASE ACTION | | Descriptor: | BETA2-BUNGAROTOXIN, SODIUM ION | | Authors: | Kwong, P.D, Mcdonald, N.Q, Sigler, P.B, Hendrickson, W.A. | | Deposit date: | 1995-10-15 | | Release date: | 1996-04-03 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of beta 2-bungarotoxin: potassium channel binding by Kunitz modules and targeted phospholipase action.
Structure, 3, 1995
|
|
1Z05
 
 | | Crystal structure of the ROK family transcriptional regulator, homolog of E.coli MLC protein. | | Descriptor: | BETA-MERCAPTOETHANOL, GLYCEROL, SULFATE ION, ... | | Authors: | Minasov, G, Brunzelle, J.S, Shuvalova, L, Collart, F.R, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-03-01 | | Release date: | 2005-03-08 | | Last modified: | 2018-06-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the ROK family transcriptional regulator, homolog of E.coli MLC protein.
To be Published
|
|
1BDO
 
 | |
1HR3
 
 | |
7KU4
 
 | |
1UMU
 
 | |
1CNT
 
 | | CILIARY NEUROTROPHIC FACTOR | | Descriptor: | CILIARY NEUROTROPHIC FACTOR, SULFATE ION, YTTERBIUM (III) ION | | Authors: | Mcdonald, N.Q, Panayotatos, N, Hendrickson, W.A. | | Deposit date: | 1996-06-06 | | Release date: | 1997-03-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of dimeric human ciliary neurotrophic factor determined by MAD phasing.
EMBO J., 14, 1995
|
|
2PR1
 
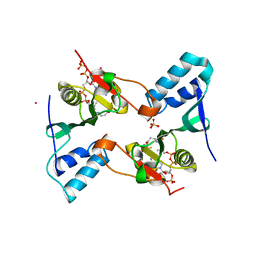 | | Crystal structure of the Bacillus subtilis N-acetyltransferase YlbP protein in complex with Coenzyme-A | | Descriptor: | COBALT (II) ION, COENZYME A, SULFATE ION, ... | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Vorontsov, I.I, Collart, F.R, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-05-03 | | Release date: | 2007-05-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of the Bacillus subtilis N-acetyltransferase YlbP protein in complex with Coenzyme-A.
To be Published
|
|
2ADI
 
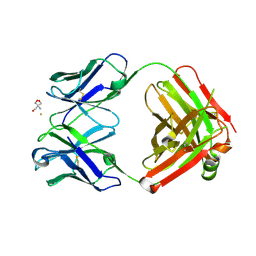 | | Crystal structure of monoclonal anti-CD4 antibody Q425 in complex with Barium | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BARIUM ION, Q425 Fab Heavy chain, ... | | Authors: | Zhou, T, Hamer, D.H, Hendrickson, W.A, Sattentau, Q.J, Kwong, P.D. | | Deposit date: | 2005-07-20 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Interfacial metal and antibody recognition.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
