4G1X
 
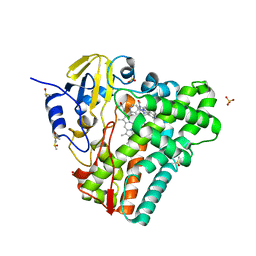 | | Crystal structure of Mycobacterium tuberculosis CYP121 in complex with 4-(1H-1,2,4-triazol-1-yl)quinolin-6-amine | | Descriptor: | 4-(1H-1,2,4-triazol-1-yl)quinolin-6-amine, Cytochrome P450 121, DIMETHYL SULFOXIDE, ... | | Authors: | Hudson, S.A. | | Deposit date: | 2012-07-11 | | Release date: | 2012-09-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Application of Fragment Screening and Merging to the Discovery of Inhibitors of the Mycobacterium tuberculosis Cytochrome P450 CYP121
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
4G48
 
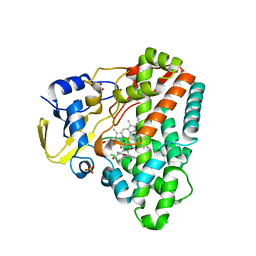 | | Structure of CYP121 in complex with 4-(4-phenoxy-1H-pyrazol-3-yl)benzene-1,3-diol | | Descriptor: | 4-(4-phenoxy-1H-pyrazol-3-yl)benzene-1,3-diol, Cytochrome P450 121, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Hudson, S.A, McLean, K.J, Surade, S, Yang, Y.-Q, Leys, D, Ciulli, A, Munro, A.W, Abell, C. | | Deposit date: | 2012-07-16 | | Release date: | 2012-09-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Application of Fragment Screening and Merging to the Discovery of Inhibitors of the Mycobacterium tuberculosis Cytochrome P450 CYP121
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
5OGX
 
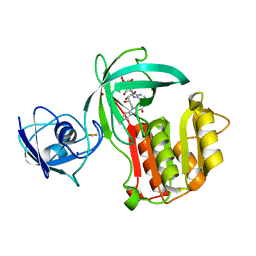 | | Crystal structure of Amycolatopsis cytochrome P450 reductase GcoB. | | Descriptor: | BROMIDE ION, Cytochrome P450 reductase, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Mallinson, S.J.B, Johnson, C.W, Neidle, E.L, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2017-07-13 | | Release date: | 2018-07-04 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | A promiscuous cytochrome P450 aromatic O-demethylase for lignin bioconversion.
Nat Commun, 9, 2018
|
|
3DYB
 
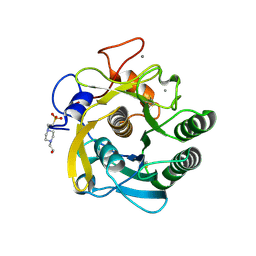 | | proteinase K- digalacturonic acid complex | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, Proteinase K, ... | | Authors: | Larson, S.B, Day, J.S, McPherson, A, Cudney, R, Nguyen, C, Center for High-Throughput Structural Biology (CHTSB) | | Deposit date: | 2008-07-25 | | Release date: | 2008-10-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | High-resolution structure of proteinase K cocrystallized with digalacturonic acid.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
8KCN
 
 | |
2N24
 
 | |
1HX6
 
 | | P3, THE MAJOR COAT PROTEIN OF THE LIPID-CONTAINING BACTERIOPHAGE PRD1. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, MAJOR CAPSID PROTEIN, ... | | Authors: | Benson, S.D, Bamford, J.K.H, Bamford, D.H, Burnett, R.M. | | Deposit date: | 2001-01-11 | | Release date: | 2001-01-24 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The X-ray crystal structure of P3, the major coat protein of the lipid-containing bacteriophage PRD1, at 1.65 A resolution.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
8JBO
 
 | | Crystal structure of TxGH116 from Thermoanaerobacterium xylanolyticum with isofagomine | | Descriptor: | 1,2-ETHANEDIOL, 5-HYDROXYMETHYL-3,4-DIHYDROXYPIPERIDINE, CALCIUM ION, ... | | Authors: | Pengthaisong, S, Ketudat Cairns, J.R. | | Deposit date: | 2023-05-09 | | Release date: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for inhibition of a GH116 beta-glucosidase and its missense mutants by GBA2 inhibitors: Crystallographic and quantum chemical study.
Chem.Biol.Interact., 384, 2023
|
|
3JW1
 
 | | Crystal Structure of Bovine Pancreatic Ribonuclease Complexed with Uridine-5'-monophosphate at 1.60 A Resolution | | Descriptor: | Ribonuclease pancreatic, URIDINE-5'-MONOPHOSPHATE | | Authors: | Larson, S.B, Day, J.S, Nguyen, C, Cudney, R, Mcpherson, A, Center for High-Throughput Structural Biology (CHTSB) | | Deposit date: | 2009-09-17 | | Release date: | 2009-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of bovine pancreatic ribonuclease complexed with uridine 5'-monophosphate at 1.60 A resolution.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
5OMU
 
 | | Crystal structure of Amycolatopsis cytochrome P450 GcoA in complex with syringol | | Descriptor: | 2,6-dimethoxyphenol, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Mallinson, S.J.B, Johnson, C.W, Neidle, E.L, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2017-08-01 | | Release date: | 2018-07-04 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A promiscuous cytochrome P450 aromatic O-demethylase for lignin bioconversion.
Nat Commun, 9, 2018
|
|
2ARC
 
 | |
2ARA
 
 | |
5OMS
 
 | | Crystal structure of Amycolatopsis cytochrome P450 GcoA in complex with guaethol. | | Descriptor: | 2-ethoxyphenol, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Mallinson, S.J.B, Johnson, C.W, Neidle, E.L, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2017-08-01 | | Release date: | 2018-07-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A promiscuous cytochrome P450 aromatic O-demethylase for lignin bioconversion.
Nat Commun, 9, 2018
|
|
5X0N
 
 | |
5X0O
 
 | |
2I5F
 
 | | Crystal structure of the C-terminal PH domain of pleckstrin in complex with D-myo-Ins(1,2,3,5,6)P5 | | Descriptor: | (1R,2R,3R,4R,5S,6S)-6-HYDROXYCYCLOHEXANE-1,2,3,4,5-PENTAYL PENTAKIS[DIHYDROGEN (PHOSPHATE)], Pleckstrin | | Authors: | Jackson, S.G, Haslam, R.J, Junop, M.S. | | Deposit date: | 2006-08-24 | | Release date: | 2007-08-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural analysis of the carboxy terminal PH domain of pleckstrin bound to D-myo-inositol 1,2,3,5,6-pentakisphosphate.
Bmc Struct.Biol., 7, 2007
|
|
1SZJ
 
 | |
2I5C
 
 | | Crystal structure of the C-terminal PH domain of pleckstrin in complex with D-myo-Ins(1,2,3,4,5)P5 | | Descriptor: | (1R,2S,3R,4S,5S,6R)-6-HYDROXYCYCLOHEXANE-1,2,3,4,5-PENTAYL PENTAKIS[DIHYDROGEN (PHOSPHATE)], Pleckstrin | | Authors: | Jackson, S.G, Haslam, R.J, Junop, M.S. | | Deposit date: | 2006-08-24 | | Release date: | 2007-08-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural analysis of the carboxy terminal PH domain of pleckstrin bound to D-myo-inositol 1,2,3,5,6-pentakisphosphate.
Bmc Struct.Biol., 7, 2007
|
|
4G47
 
 | | Structure of cytochrome P450 CYP121 in complex with 4-(1H-1,2,4-triazol-1-yl)phenol | | Descriptor: | 4-(1H-1,2,4-triazol-1-yl)phenol, Cytochrome P450 121, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Hudson, S.A, McLean, K.J, Surade, S, Yang, Y.-Q, Leys, D, Ciulli, A, Munro, A.W, Abell, C. | | Deposit date: | 2012-07-16 | | Release date: | 2012-09-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Application of Fragment Screening and Merging to the Discovery of Inhibitors of the Mycobacterium tuberculosis Cytochrome P450 CYP121
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
4G45
 
 | | Structure of cytochrome CYP121 in complex with 2-methylquinolin-6-amine | | Descriptor: | 2-methylquinolin-6-amine, Cytochrome P450 121, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Hudson, S.A, McLean, K.J, Surade, S, Yang, Y.-Q, Leys, D, Ciulli, A, Munro, A.W, Abell, C. | | Deposit date: | 2012-07-16 | | Release date: | 2012-09-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Application of Fragment Screening and Merging to the Discovery of Inhibitors of the Mycobacterium tuberculosis Cytochrome P450 CYP121
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
4G46
 
 | | Structure of cytochrome P450 CYP121 in complex with 4-oxo-4,5,6,7-tetrahydrobenzofuran-3-carboxylate | | Descriptor: | 4-oxo-4,5,6,7-tetrahydro-1-benzofuran-3-carboxylic acid, Cytochrome P450 121, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Hudson, S.A, McLean, K.J, Surade, S, Yang, Y.-Q, Leys, D, Ciulli, A, Munro, A.W, Abell, C. | | Deposit date: | 2012-07-16 | | Release date: | 2012-09-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Application of Fragment Screening and Merging to the Discovery of Inhibitors of the Mycobacterium tuberculosis Cytochrome P450 CYP121
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
4G44
 
 | | Structure of P450 CYP121 in complex with lead compound MB286, 3-((1H-1,2,4-triazol-1-yl)methyl)aniline | | Descriptor: | 3-(1H-1,2,4-triazol-1-ylmethyl)aniline, Cytochrome P450 121, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Hudson, S.A, McLean, K.J, Surade, S, Yang, Y.-Q, Leys, D, Ciulli, A, Munro, A.W, Abell, C. | | Deposit date: | 2012-07-16 | | Release date: | 2012-09-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Application of Fragment Screening and Merging to the Discovery of Inhibitors of the Mycobacterium tuberculosis Cytochrome P450 CYP121
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
3ENJ
 
 | | Structure of Pig Heart Citrate Synthase at 1.78 A resolution | | Descriptor: | CHLORIDE ION, CYSTEINE, Citrate synthase, ... | | Authors: | Larson, S.B, Day, J.S, Nguyen, C, Cudney, R, McPherson, A, Center for High-Throughput Structural Biology (CHTSB) | | Deposit date: | 2008-09-25 | | Release date: | 2009-02-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure of pig heart citrate synthase at 1.78 A resolution.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
1CJD
 
 | | THE BACTERIOPHAGE PRD1 COAT PROTEIN, P3, IS STRUCTURALLY SIMILAR TO HUMAN ADENOVIRUS HEXON | | Descriptor: | PROTEIN (MAJOR CAPSID PROTEIN (P3)) | | Authors: | Benson, S.D, Bamford, J.K.H, Bamford, D.H, Burnett, R.M. | | Deposit date: | 1999-04-12 | | Release date: | 1999-09-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Viral evolution revealed by bacteriophage PRD1 and human adenovirus coat protein structures.
Cell(Cambridge,Mass.), 98, 1999
|
|
1U8O
 
 | | Crystal structure of the HIV-1 Cross Neutralizing Monoclonal Antibody 2F5 in complex with gp41 Peptide ELDKHAS | | Descriptor: | ANTIBODY 2F5 (HEAVY CHAIN), ANTIBODY 2F5 (LIGHT CHAIN), GP41 PEPTIDE | | Authors: | Bryson, S, Julien, J.-P, Hynes, R.C, Pai, E.F. | | Deposit date: | 2004-08-06 | | Release date: | 2004-10-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Crystallographic definition of the epitope promiscuity of the broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2F5: vaccine design implications
J.Virol., 83, 2009
|
|
