1JUF
 
 | | Structure of Minor Histocompatibility Antigen peptide, H13b, complexed to H2-Db | | Descriptor: | Beta-2-microglobulin, H13b peptide, H2-Db major histocompatibility antigen | | Authors: | Ostrov, D.A, Roden, M.M, Shi, W, Palmieri, E, Christianson, G.J, Mendoza, L, Villaflor, G, Tilley, D, Shastri, N, Grey, H, Almo, S.C, Roopenian, D.C, Nathenson, S.G. | | Deposit date: | 2001-08-24 | | Release date: | 2002-03-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | How H13 histocompatibility peptides differing by a single methyl group and lacking conventional MHC binding anchor motifs determine self-nonself discrimination.
J.Immunol., 168, 2002
|
|
1RPE
 
 | | THE PHAGE 434 OR2/R1-69 COMPLEX AT 2.5 ANGSTROMS RESOLUTION | | Descriptor: | DNA (5'-D(*AP*CP*AP*AP*AP*CP*AP*AP*GP*AP*TP*AP*CP*AP*TP*TP*G P*TP*AP*T)-3'), DNA (5'-D(*TP*AP*TP*AP*CP*AP*AP*TP*GP*TP*AP*TP*CP*TP*TP*GP*T P*TP*TP*G)-3'), PROTEIN (434 REPRESSOR) | | Authors: | Shimon, L.J.W, Harrison, S.C. | | Deposit date: | 1993-03-24 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The phage 434 OR2/R1-69 complex at 2.5 A resolution.
J.Mol.Biol., 232, 1993
|
|
1C8C
 
 | | CRYSTAL STRUCTURES OF THE CHROMOSOMAL PROTEINS SSO7D/SAC7D BOUND TO DNA CONTAINING T-G MISMATCHED BASE PAIRS | | Descriptor: | 5'-D(*GP*TP*GP*AP*TP*CP*GP*C)-3', DNA-BINDING PROTEIN 7A | | Authors: | Su, S, Gao, Y.-G, Robinson, H, Liaw, Y.-C, Edmondson, S.P, Shriver, J.W, Wang, A.H.-J. | | Deposit date: | 2000-05-04 | | Release date: | 2001-05-04 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structures of the chromosomal proteins Sso7d/Sac7d bound to DNA containing T-G mismatched base-pairs.
J.Mol.Biol., 303, 2000
|
|
2IUH
 
 | | Crystal structure of the PI3-kinase p85 N-terminal SH2 domain in complex with c-Kit phosphotyrosyl peptide | | Descriptor: | C-KIT PHOSPHOTYROSYL PEPTIDE, PHOSPHATIDYLINOSITOL 3-KINASE REGULATORY ALPHA SUBUNIT | | Authors: | Nolte, R.T, Eck, M.J, Schlessinger, J, Shoelson, S.E, Harrison, S.C. | | Deposit date: | 2006-06-03 | | Release date: | 2006-06-06 | | Last modified: | 2021-04-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Pi 3-Kinase P85 Amino-Terminal Sh2 Domain and its Phosphopeptide Complexes
Nat.Struct.Biol., 3, 1996
|
|
2IUG
 
 | | Crystal structure of the PI3-kinase p85 N-terminal SH2 domain | | Descriptor: | PHOSPHATIDYLINOSITOL 3-KINASE REGULATORY ALPHA SUBUNIT | | Authors: | Nolte, R.T, Eck, M.J, Schlessinger, J, Shoelson, S.E, Harrison, S.C. | | Deposit date: | 2006-06-03 | | Release date: | 2006-06-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal Structure of the Pi 3-Kinase P85 Amino-Terminal Sh2 Domain and its Phosphopeptide Complexes
Nat.Struct.Biol., 3, 1996
|
|
2IUI
 
 | | Crystal structure of the PI3-kinase p85 N-terminal SH2 domain in complex with PDGFR phosphotyrosyl peptide | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Platelet-derived growth factor receptor beta | | Authors: | Nolte, R.T, Eck, M.J, Schlessinger, J, Shoelson, S.E, Harrison, S.C. | | Deposit date: | 2006-06-03 | | Release date: | 2006-06-06 | | Last modified: | 2021-04-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the Pi 3-Kinase P85 Amino- Terminal Sh2 Domain and its Phosphopeptide Complexes
Nat.Struct.Biol., 3, 1996
|
|
4V2K
 
 | | Crystal structure of the thiosulfate dehydrogenase TsdA in complex with thiosulfate | | Descriptor: | HEME C, THIOSULFATE, THIOSULFATE DEHYDROGENASE | | Authors: | Grabarczyk, D.B, Chappell, P.E, Eisel, B, Johnson, S, Lea, S.M, Berks, B.C. | | Deposit date: | 2014-10-10 | | Release date: | 2015-02-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Mechanism of Thiosulfate Oxidation in the Soxa Family of Cysteine-Ligated Cytochromes
J.Biol.Chem., 290, 2015
|
|
4UWQ
 
 | | Crystal structure of the disulfide-linked complex of the thiosulfodyrolase SoxB with the carrier-protein SoxYZ from Thermus thermophilus | | Descriptor: | MANGANESE (II) ION, SOXY PROTEIN, SOXZ, ... | | Authors: | Grabarczyk, D.B, Chappell, P.E, Johnson, S, Stelzl, L.S, Lea, S.M, Berks, B.C. | | Deposit date: | 2014-08-14 | | Release date: | 2015-12-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.28 Å) | | Cite: | Structural Basis for Specificity and Promiscuity in a Carrier Protein/Enzyme System from the Sulfur Cycle
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5UC0
 
 | | Crystal Structure of Beta-barrel-like, Uncharacterized Protein of COG5400 from Brucella abortus | | Descriptor: | CHLORIDE ION, PENTAETHYLENE GLYCOL, SULFATE ION, ... | | Authors: | Kim, Y, Bigelow, L, Endres, M, Babnigg, G, Crosson, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2016-12-21 | | Release date: | 2017-02-08 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Periplasmic protein EipA determines envelope stress resistance and virulence in Brucella abortus.
Mol. Microbiol., 2018
|
|
3F5K
 
 | | Semi-active E176Q mutant of rice BGlu1, a plant exoglucanase/beta-glucosidase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-glucosidase, GLYCEROL, ... | | Authors: | Chuenchor, W, Ketudat Cairns, J.R, Pengthaisong, S, Robinson, R.C, Yuvaniyama, J, Chen, C.-J. | | Deposit date: | 2008-11-03 | | Release date: | 2009-11-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structural basis of oligosaccharide binding by rice BGlu1 beta-glucosidase
J.Struct.Biol., 173, 2011
|
|
2P66
 
 | | Human DNA Polymerase beta complexed with tetrahydrofuran (abasic site) containing DNA | | Descriptor: | DNA (5'-D(*CP*CP*GP*AP*CP*AP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*CP*C)-3'), DNA (5'-D(P*(3DR)P*GP*TP*CP*GP*G)-3'), ... | | Authors: | Prasad, R, Batra, V.K, Yang, X.-P, Krahn, J.M, Pedersen, L.C, Beard, W.A, Wilson, S.H. | | Deposit date: | 2007-03-16 | | Release date: | 2007-04-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insight into the DNA Polymerase beta deoxyribose phosphate lyase mechanism
DNA REPAIR, 4, 2005
|
|
3F64
 
 | | F17a-G lectin domain with bound GlcNAc(beta1-O)paranitrophenyl ligand | | Descriptor: | 4-nitrophenyl 2-acetamido-2-deoxy-beta-D-glucopyranoside, F17a-G | | Authors: | Buts, L, De Boer, A, Olsson, J.D.M, Jonckheere, W, De Kerpel, M, De Genst, E, Guerardel, Y, Willaert, R, Wyns, L, Wuhrer, M, Oscarson, S, De Greve, H, Bouckaert, J. | | Deposit date: | 2008-11-05 | | Release date: | 2009-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Sampling of Glycan Interaction Profiles Reveals Mucosal Receptors for Fimbrial Adhesins of Enterotoxigenic Escherichia coli.
Biology (Basel), 2, 2013
|
|
2DSH
 
 | | Crystal structure of Lys26 to Tyr mutant of Diphthine synthase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Mizutani, H, Matsuura, Y, Saraboji, K, Malathy Sony, S.M, Ponnuswamy, M.N, Kumarevel, T.S, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-06-30 | | Release date: | 2006-12-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of diphthine synthase from Pyrococcus horikoshii OT3
To be Published
|
|
2P8L
 
 | | Crystal structure of the HIV-1 Cross Neutralizing Monoclonal Antibody 2F5 in complex with gp41 Peptide ELLELDKWASLWN | | Descriptor: | gp41 peptide, nmAb 2F5, heavy chain, ... | | Authors: | Julien, J.P, Bryson, S, Pai, E.F. | | Deposit date: | 2007-03-22 | | Release date: | 2007-05-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Structural details of HIV-1 recognition by the broadly neutralizing monoclonal antibody 2F5: epitope conformation, antigen-recognition loop mobility, and anion-binding site.
J.Mol.Biol., 384, 2008
|
|
2LHR
 
 | |
3F5J
 
 | | Semi-active E176Q mutant of rice BGlu1, a plant exoglucanase/beta-glucosidase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-glucosidase, SULFATE ION, ... | | Authors: | Chuenchor, W, Ketudat Cairns, J.R, Pengthaisong, S, Robinson, R.C, Yuvaniyama, J, Chen, C.-J. | | Deposit date: | 2008-11-03 | | Release date: | 2009-11-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The structural basis of oligosaccharide binding by rice BGlu1 beta-glucosidase
J.Struct.Biol., 173, 2011
|
|
2DSI
 
 | | Crystal structure of Glu171 to Arg mutant of Diphthine synthase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Mizutani, H, Matsuura, Y, Saraboji, K, Malathy Sony, S.M, Ponnuswamy, M.N, Kumarevel, T.S, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-06-30 | | Release date: | 2006-12-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of diphthine synthase from Pyrococcus horikoshii OT3
To be Published
|
|
2DSG
 
 | | Crystal structure of Lys26 to Arg mutant of Diphthine synthase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Mizutani, H, Matsuura, Y, Saraboji, K, Malathy Sony, S.M, Ponnuswamy, M.N, Kumarevel, T.S, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-06-30 | | Release date: | 2006-12-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of diphthine synthase from Pyrococcus horikoshii OT3
To be Published
|
|
1FN9
 
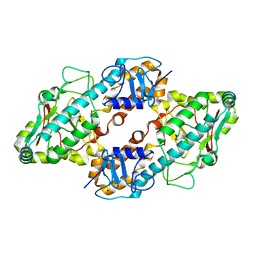 | | CRYSTAL STRUCTURE OF THE REOVIRUS OUTER CAPSID PROTEIN SIGMA 3 | | Descriptor: | OUTER-CAPSID PROTEIN SIGMA 3, ZINC ION | | Authors: | Olland, A.M, Jane-Valbuena, J, Schiff, L.A, Nibert, M.L, Harrison, S.C. | | Deposit date: | 2000-08-21 | | Release date: | 2001-03-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the reovirus outer capsid and dsRNA-binding protein sigma3 at 1.8 A resolution.
EMBO J., 20, 2001
|
|
2DV7
 
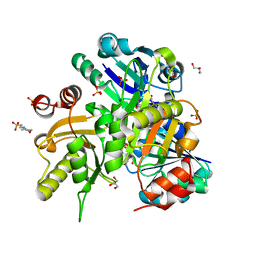 | | Crystal structure of Lys187 to Arg mutant of Diphthine synthase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Mizutani, H, Matsuura, Y, Saraboji, K, Malathy Sony, S.M, Ponnuswamy, M.N, Kumarevel, T.S, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-07-28 | | Release date: | 2007-01-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of diphthine synthase from Pyrococcus horikoshii OT3
To be Published
|
|
8FNW
 
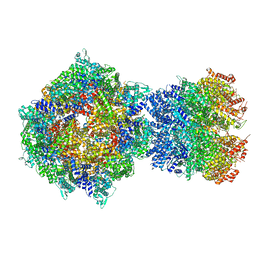 | | Structure of RdrA-RdrB complex from Escherichia coli RADAR defense system | | Descriptor: | Adenosine deaminase, Archaeal ATPase, ZINC ION | | Authors: | Duncan-Lowey, B, Johnson, A.G, Rawson, S, Mayer, M.L, Kranzusch, P.J. | | Deposit date: | 2022-12-28 | | Release date: | 2023-02-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (6.73 Å) | | Cite: | Cryo-EM structure of the RADAR supramolecular anti-phage defense complex.
Cell, 186, 2023
|
|
8FNV
 
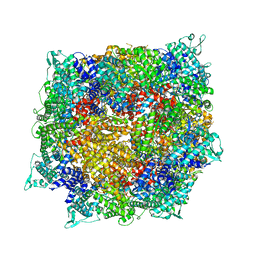 | | Structure of RdrB from Escherichia coli RADAR defense system | | Descriptor: | Adenosine deaminase, ZINC ION | | Authors: | Duncan-Lowey, B, Johnson, A.G, Rawson, S, Mayer, M.L, Kranzusch, P.J. | | Deposit date: | 2022-12-28 | | Release date: | 2023-02-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.11 Å) | | Cite: | Cryo-EM structure of the RADAR supramolecular anti-phage defense complex.
Cell, 186, 2023
|
|
2RAK
 
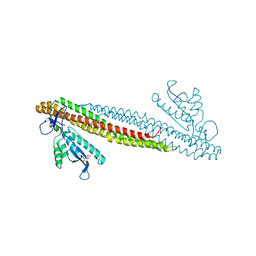 | | PI(3)P bound PX-BAR membrane remodeling unit of Sorting Nexin 9 | | Descriptor: | 2-(BUTANOYLOXY)-1-{[(HYDROXY{[2,3,4,6-TETRAHYDROXY-5-(PHOSPHONOOXY)CYCLOHEXYL]OXY}PHOSPHORYL)OXY]METHYL}ETHYL BUTANOATE, Sorting nexin-9 | | Authors: | Pylypenko, O, Lundmark, R, Rasmuson, E, Carlsson, S.R, Rak, A. | | Deposit date: | 2007-09-16 | | Release date: | 2007-12-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The PX-BAR membrane-remodeling unit of sorting nexin 9
Embo J., 26, 2007
|
|
1N7U
 
 | | THE RECEPTOR-BINDING PROTEIN P2 OF BACTERIOPHAGE PRD1: CRYSTAL FORM I | | Descriptor: | ACETATE ION, Adsorption protein P2, CALCIUM ION | | Authors: | Xu, L, Benson, S.D, Butcher, S.J, Bamford, D.H, Burnett, R.M. | | Deposit date: | 2002-11-18 | | Release date: | 2003-04-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Receptor Binding Protein P2 of PRD1, a
Virus Targeting Antibiotic-Resistant Bacteria,
Has a Novel Fold Suggesting Multiple Functions.
Structure, 11, 2003
|
|
1NCN
 
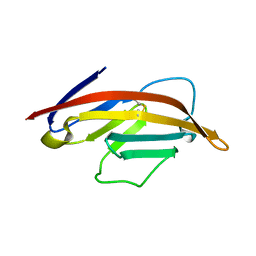 | |
