4RL5
 
 | |
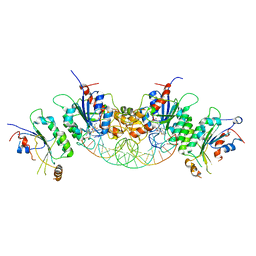
6BRR
 
 | |
6CWZ
 
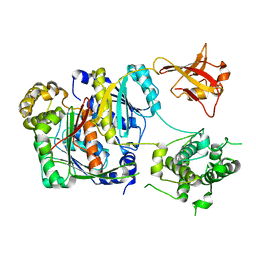 | | Crystal structure of apo SUMO E1 | | Descriptor: | SUMO-activating enzyme subunit 1, SUMO-activating enzyme subunit 2, ZINC ION | | Authors: | Lv, Z, Yuan, L, Atkison, J.H, Williams, K.M, Olsen, S.K. | | Deposit date: | 2018-04-01 | | Release date: | 2019-01-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Molecular mechanism of a covalent allosteric inhibitor of SUMO E1 activating enzyme.
Nat Commun, 9, 2018
|
|
7MLQ
 
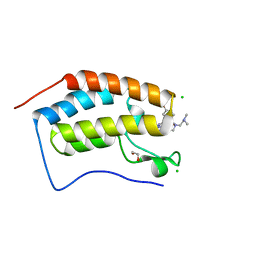 | | X-ray crystal structure of human BRD4(D1) in complex with 2-(4-{5-[6-(2,5-dibromophenoxy)pyridin-2-yl]-4-methyl-1H-1,2,3-triazol-1-yl}piperidin-1-yl)-N,N-dimethylethan-1-amine (compound 26) | | Descriptor: | 1,2-ETHANEDIOL, 2-(4-{5-[6-(2,5-dibromophenoxy)pyridin-2-yl]-4-methyl-1H-1,2,3-triazol-1-yl}piperidin-1-yl)-N,N-dimethylethan-1-amine, Bromodomain-containing protein 4, ... | | Authors: | Cui, H, Johnson, J.A, Vail, N.R, Shi, K, Aihara, H, Pomerantz, W.C.K. | | Deposit date: | 2021-04-28 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | 4-Methyl-1,2,3-Triazoles as N -Acetyl-Lysine Mimics Afford Potent BET Bromodomain Inhibitors with Improved Selectivity.
J.Med.Chem., 64, 2021
|
|
7MLS
 
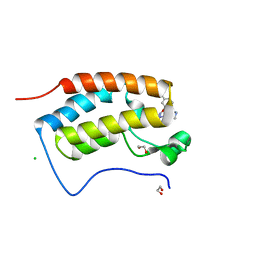 | | X-ray crystal structure of human BRD4(D1) in complex with 2-(2,5-dibromophenoxy)-6-[4-methyl-1-(piperidin-4-yl)-1H-1,2,3-triazol-5-yl]pyridine (compound 23) | | Descriptor: | 1,2-ETHANEDIOL, 2-(2,5-dibromophenoxy)-6-[4-methyl-1-(piperidin-4-yl)-1H-1,2,3-triazol-5-yl]pyridine, Bromodomain-containing protein 4, ... | | Authors: | Cui, H, Johnson, J.A, Zahid, H, Buchholz, C.R, Shi, K, Aihara, H, Pomerantz, W.C.K. | | Deposit date: | 2021-04-28 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | 4-Methyl-1,2,3-Triazoles as N -Acetyl-Lysine Mimics Afford Potent BET Bromodomain Inhibitors with Improved Selectivity.
J.Med.Chem., 64, 2021
|
|
1XI3
 
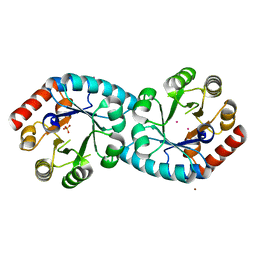 | | Thiamine phosphate pyrophosphorylase from Pyrococcus furiosus Pfu-1255191-001 | | Descriptor: | NICKEL (II) ION, SULFATE ION, Thiamine phosphate pyrophosphorylase, ... | | Authors: | Zhou, W, Chen, L, Tempel, W, Liu, Z.-J, Habel, J, Lee, D, Lin, D, Chang, S.-H, Dillard, B.D, Eneh, J.C, Hopkins, R.C, Jenney Jr, F.E, Kelley, L.-L.C, Lee, H.-S, Li, T, Poole II, F.L, Shah, C, Sugar, F.J, Arendall III, W.B, Richardson, J.S, Richardson, D.C, Rose, J.P, Adams, M.W.W, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-09-21 | | Release date: | 2004-09-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Thiamine phosphate pyrophosphorylase from Pyrococcus furiosus Pfu-1255191-001
To be published
|
|
1XI9
 
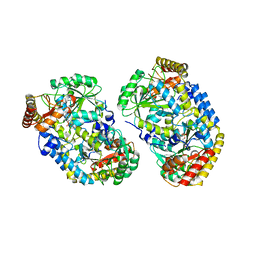 | | Alanine aminotransferase from Pyrococcus furiosus Pfu-1397077-001 | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, putative transaminase | | Authors: | Zhou, W, Tempel, W, Shah, A, Chen, L, Liu, Z.-J, Lee, D, Lin, D, Chang, S.-H, Brereton, P.S, Izumi, M, Jenney Jr, F.E, Lee, H.-S, Poole II, F.L, Shah, C, Sugar, F.J, Arendall III, W.B, Richardson, J.S, Richardson, D.C, Rose, J.P, Adams, M.W.W, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-09-21 | | Release date: | 2004-11-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Alanine aminotransferase from Pyrococcus furiosus Pfu-1397077-001
To be published
|
|
1IKG
 
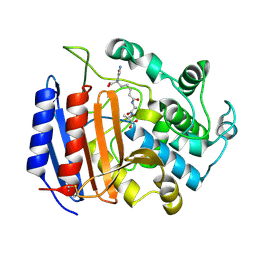 | | MICHAELIS COMPLEX OF STREPTOMYCES R61 DD-PEPTIDASE WITH A SPECIFIC PEPTIDOGLYCAN SUBSTRATE FRAGMENT | | Descriptor: | D-ALANYL-D-ALANINE CARBOXYPEPTIDASE, GLYCYL-L-ALPHA-AMINO-EPSILON-PIMELYL-D-ALANYL-D-ALANINE | | Authors: | Mcdonough, M.A, Anderson, J.W, Silvaggi, N.R, Pratt, R.F, Knox, J.R, Kelly, J.A. | | Deposit date: | 2001-05-03 | | Release date: | 2002-09-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of two kinetic intermediates reveal species specificity of penicillin-binding proteins.
J.Mol.Biol., 322, 2002
|
|
3A29
 
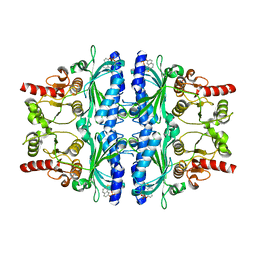 | | Crystal structure of human liver FBPase in complex with tricyclic inhibitor | | Descriptor: | 2-amino-4,5-dihydronaphtho[1,2-d][1,3]thiazol-8-yl dihydrogen phosphate, Fructose-1,6-bisphosphatase 1 | | Authors: | Takahashi, M, Sone, J, Hanzawa, H. | | Deposit date: | 2009-05-08 | | Release date: | 2009-10-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Synthesis, SAR, and X-ray structure of tricyclic compounds as potent FBPase inhibitors
Bioorg.Med.Chem.Lett., 19, 2009
|
|
1S1K
 
 | |
3LR8
 
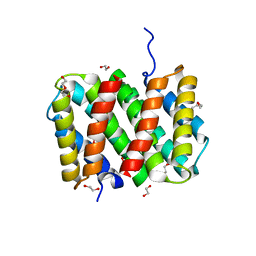 | | Self-assembly of spider silk proteins is controlled by a pH-sensitive relay | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Major ampullate spidroin 1, ... | | Authors: | Askarieh, G, Hedhammar, H, Nordling, K, Rising, A, Johansson, J, Knight, S.D. | | Deposit date: | 2010-02-10 | | Release date: | 2010-05-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Self-assembly of spider silk proteins is controlled by a pH-sensitive relay.
Nature, 465, 2010
|
|
3GL6
 
 | |
3H57
 
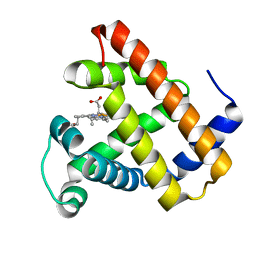 | | Myoglobin Cavity Mutant H64LV68N Deoxy form | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Soman, J, Olson, J.S. | | Deposit date: | 2009-04-21 | | Release date: | 2009-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Optical detection of disordered water within a protein cavity.
J.Am.Chem.Soc., 131, 2009
|
|
4OXI
 
 | | Crystal structure of Vibrio cholerae adenylation domain AlmE in complex with glycyl-adenosine-5'-phosphate | | Descriptor: | Enterobactin synthetase component F-related protein, GLYCYL-ADENOSINE-5'-PHOSPHATE | | Authors: | Fage, C.D, Henderson, J.C, Keatinge-Clay, A.T, Trent, M.S. | | Deposit date: | 2014-02-05 | | Release date: | 2014-12-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.261 Å) | | Cite: | Antimicrobial peptide resistance of Vibrio cholerae results from an LPS modification pathway related to nonribosomal peptide synthetases.
Acs Chem.Biol., 9, 2014
|
|
1AJU
 
 | |
1IF0
 
 | | PSEUDO-ATOMIC MODEL OF BACTERIOPHAGE HK97 PROCAPSID (PROHEAD II) | | Descriptor: | PROTEIN (MAJOR CAPSID PROTEIN GP5) | | Authors: | Conway, J.F, Wikoff, W.R, Cheng, N, Duda, R.L, Hendrix, R.W, Johnson, J.E, Steven, A.C. | | Deposit date: | 2001-04-11 | | Release date: | 2001-05-02 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Virus maturation involving large subunit rotations and local refolding.
Science, 292, 2001
|
|
1F2N
 
 | | RICE YELLOW MOTTLE VIRUS | | Descriptor: | CALCIUM ION, CAPSID PROTEIN | | Authors: | Qu, C, Liljas, L, Opalka, N, Brugidou, C, Yeager, M, Beachy, R.N, Fauquet, C.M, Johnson, J.E, Lin, T. | | Deposit date: | 2000-05-26 | | Release date: | 2000-11-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | 3D domain swapping modulates the stability of members of an icosahedral virus group.
Structure Fold.Des., 8, 2000
|
|
1RYQ
 
 | | Putative DNA-directed RNA polymerase, subunit e'' from Pyrococcus Furiosus Pfu-263306-001 | | Descriptor: | DNA-directed RNA polymerase, subunit e'', ZINC ION | | Authors: | Liu, Z.-J, Chen, L, Tempel, W, Shah, A, Arendall III, W.B, Rose, J.P, Brereton, P.S, Izumi, M, Jenney Jr, F.E, Lee, H.S, Poole II, F.L, Shah, C, Sugar, F.J, Adams, M.W.W, Richardson, D.C, Richardson, J.S, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2003-12-22 | | Release date: | 2004-08-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Parameter-space screening: a powerful tool for high-throughput crystal structure determination.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
4LKM
 
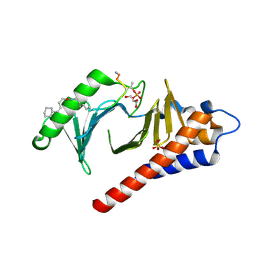 | | Crystal structure of Plk1 Polo-box domain in complex with PL-74 | | Descriptor: | GLYCEROL, PL-74, SULFATE ION, ... | | Authors: | Lee, W.C, Song, J.H, Kim, H.Y. | | Deposit date: | 2013-07-08 | | Release date: | 2013-12-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Exploring the binding nature of pyrrolidine pocket-dependent interactions in the polo-box domain of polo-like kinase 1
Plos One, 8, 2013
|
|
4ICG
 
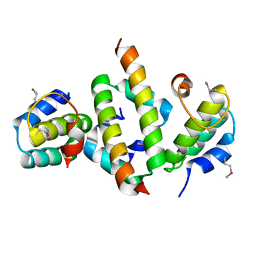 | | N-terminal dimerization domain of H-NS in complex with Hha (Salmonella Typhimurium) | | Descriptor: | DNA-binding protein H-NS, Hemolysin expression modulating protein (Involved in environmental regulation of virulence factors) | | Authors: | Ali, S.S, Whitney, J.C, Stevenson, J, Robinson, H, Howell, P.L, Navarre, W.W. | | Deposit date: | 2012-12-10 | | Release date: | 2013-03-27 | | Last modified: | 2016-05-18 | | Method: | X-RAY DIFFRACTION (2.9217 Å) | | Cite: | Structural Insights into the Regulation of Foreign Genes in Salmonella by the Hha/H-NS Complex.
J.Biol.Chem., 288, 2013
|
|
3IKK
 
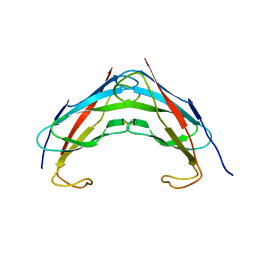 | | Crystal structure analysis of msp domain | | Descriptor: | Vesicle-associated membrane protein-associated protein B/C | | Authors: | Shi, J, Lua, S, Song, J. | | Deposit date: | 2009-08-06 | | Release date: | 2010-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Elimination of the native structure and solubility of the hVAPB MSP domain by the Pro56Ser mutation that causes amyotrophic lateral sclerosis.
Biochemistry, 49, 2010
|
|
4M83
 
 | | Ensemble refinement of protein crystal structure (2IYF) of macrolide glycosyltransferases OleD complexed with UDP and Erythromycin A | | Descriptor: | ERYTHROMYCIN A, MAGNESIUM ION, Oleandomycin glycosyltransferase, ... | | Authors: | Wang, F, Helmich, K.E, Xu, W, Singh, S, Olmos Jr, J.L, Martinez iii, E, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2013-08-12 | | Release date: | 2013-09-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | Crystal structure of macrolide glycosyltransferases OleD
To be Published
|
|
3ND0
 
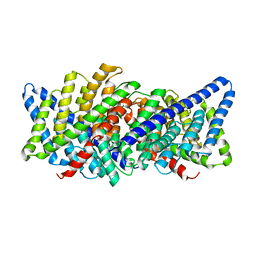 | | X-ray crystal structure of a slow cyanobacterial Cl-/H+ antiporter | | Descriptor: | CHLORIDE ION, Sll0855 protein | | Authors: | Jayaram, H, Robertson, J, Wu, F, Williams, C, Miller, C. | | Deposit date: | 2010-06-06 | | Release date: | 2011-01-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of a Slow CLC Cl(-)/H(+) Antiporter from a Cyanobacterium.
Biochemistry, 50, 2011
|
|
1NNQ
 
 | | rubrerythrin from Pyrococcus furiosus Pfu-1210814 | | Descriptor: | Rubrerythrin, ZINC ION | | Authors: | Liu, Z.-J, Tempel, W, Schubot, F.D, Shah, A, Arendall III, W.B, Rose, J.P, Richardson, D.C, Richardson, J.S, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2003-01-14 | | Release date: | 2004-03-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural genomics of Pyrococcus furiosus: X-ray crystallography reveals 3D domain swapping in rubrerythrin
Proteins, 57, 2004
|
|
1XAX
 
 | | NMR structure of HI0004, a putative essential gene product from Haemophilus influenzae | | Descriptor: | Hypothetical UPF0054 protein HI0004 | | Authors: | Yeh, D.C, Parsons, J.F, Parsons, L.M, Liu, F, Eisenstein, E, Orban, J, Structure 2 Function Project (S2F) | | Deposit date: | 2004-08-26 | | Release date: | 2005-01-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of HI0004, a putative essential gene product from Haemophilus influenzae, and comparison with the X-ray structure of an Aquifex aeolicus homolog
Protein Sci., 14, 2005
|
|
