8UF2
 
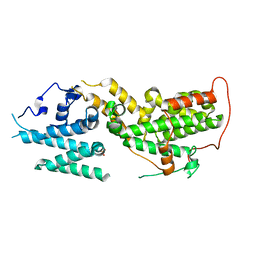 | | Apo SOS2 crystal structure in P1 space group | | Descriptor: | SULFATE ION, Son of sevenless homolog 2 | | Authors: | Gunn, R.J, Lawson, J.D. | | Deposit date: | 2023-10-03 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of Five SOS2 Fragment Hits with Binding Modes Determined by SOS2 X-Ray Cocrystallography.
J.Med.Chem., 67, 2024
|
|
8UH0
 
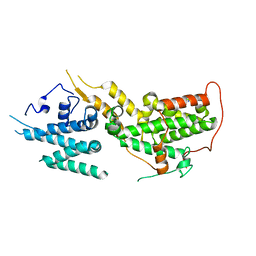 | |
8UC9
 
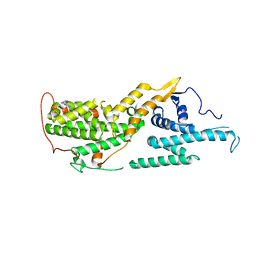 | |
1WCQ
 
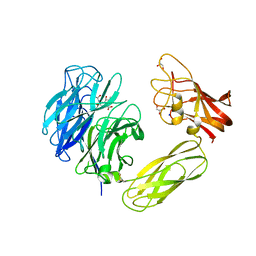 | | Mutagenesis of the Nucleophilic Tyrosine in a Bacterial Sialidase to Phenylalanine. | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, GLYCEROL, SIALIDASE, ... | | Authors: | Newstead, S, Watson, J.N, Bennet, A.J, Taylor, G. | | Deposit date: | 2004-11-19 | | Release date: | 2005-10-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Two Nucleophilic Mutants of the Micromonospora Viridifaciens Sialidase Operate with Retention of Configuration by Two Different Mechanisms.
Chembiochem, 6, 2005
|
|
2RGJ
 
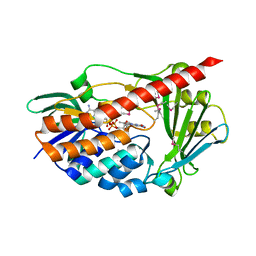 | | Crystal structure of flavin-containing monooxygenase PhzS | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Flavin-containing monooxygenase | | Authors: | Ladner, J.E, Parsons, J.F, Greenhagen, B.T, Robinson, H. | | Deposit date: | 2007-10-03 | | Release date: | 2008-05-20 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the Pyocyanin Biosynthetic Protein PhzS.
Biochemistry, 47, 2008
|
|
8V8H
 
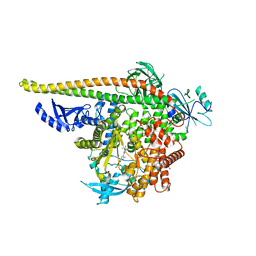 | | PI3Ka H1047R co-crystal structure with inhibitor in cryptic pocket near H1047R (compound 4). | | Descriptor: | (2S)-N~1~-[(4P)-2-tert-butyl-4'-methyl[4,5'-bi-1,3-thiazol]-2'-yl]pyrrolidine-1,2-dicarboxamide, 2-({(1R)-1-[2-(4,4-dimethylpiperidin-1-yl)-3,6-dimethyl-4-oxo-4H-1-benzopyran-8-yl]ethyl}amino)benzoic acid, Phosphatidylinositol 3-kinase regulatory subunit alpha, ... | | Authors: | Gunn, R.J, Lawson, J.D. | | Deposit date: | 2023-12-05 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (3.58 Å) | | Cite: | Discovery of Pyridopyrimidinones that Selectively Inhibit the H1047R PI3K alpha Mutant Protein.
J.Med.Chem., 67, 2024
|
|
8V8I
 
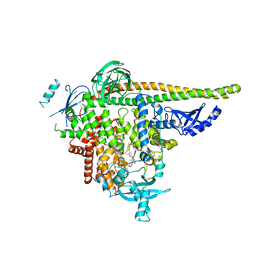 | | PI3Ka H1047R co-crystal structure with inhibitor in cryptic pocket (compound 5). | | Descriptor: | (2S)-N~1~-[(4P)-2-tert-butyl-4'-methyl[4,5'-bi-1,3-thiazol]-2'-yl]pyrrolidine-1,2-dicarboxamide, CHLORIDE ION, N-{(3S)-3-(2-methylphenyl)-6-[(oxetan-3-yl)amino]-1-oxo-2,3-dihydro-1H-isoindol-4-yl}-1-benzothiophene-3-carboxamide, ... | | Authors: | Gunn, R.J, Lawson, J.D. | | Deposit date: | 2023-12-05 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Discovery of Pyridopyrimidinones that Selectively Inhibit the H1047R PI3K alpha Mutant Protein.
J.Med.Chem., 67, 2024
|
|
8V8V
 
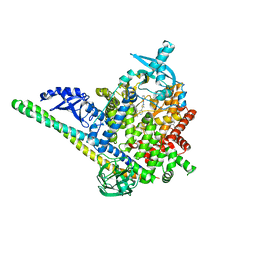 | | PI3Ka H1047R co-crystal structure with inhibitor in cryptic pocket near H1047R (compound 7). | | Descriptor: | (2S)-N~1~-{4-methyl-5-[2-(1,1,1-trifluoro-2-methylpropan-2-yl)pyridin-4-yl]-1,3-thiazol-2-yl}pyrrolidine-1,2-dicarboxamide, 2-[[(1~{R})-1-(7-methyl-4-oxidanylidene-2-piperidin-1-yl-3~{H}-pyrido[1,2-a]pyrimidin-9-yl)ethyl]amino]benzoic acid, Phosphatidylinositol 3-kinase regulatory subunit alpha, ... | | Authors: | Gunn, R.J, Lawson, J.D. | | Deposit date: | 2023-12-06 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Discovery of Pyridopyrimidinones that Selectively Inhibit the H1047R PI3K alpha Mutant Protein.
J.Med.Chem., 67, 2024
|
|
8V8J
 
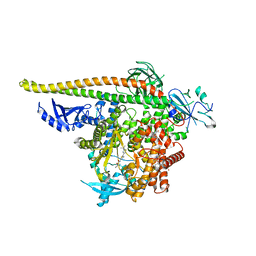 | | PI3Ka H1047R co-crystal structure with inhibitors in two cryptic pockets (compounds 4 and 5). | | Descriptor: | (2S)-N~1~-[(4P)-2-tert-butyl-4'-methyl[4,5'-bi-1,3-thiazol]-2'-yl]pyrrolidine-1,2-dicarboxamide, 2-({(1R)-1-[2-(4,4-dimethylpiperidin-1-yl)-3,6-dimethyl-4-oxo-4H-1-benzopyran-8-yl]ethyl}amino)benzoic acid, N-{(3S)-3-(2-methylphenyl)-6-[(oxetan-3-yl)amino]-1-oxo-2,3-dihydro-1H-isoindol-4-yl}-1-benzothiophene-3-carboxamide, ... | | Authors: | Gunn, R.J, Lawson, J.D. | | Deposit date: | 2023-12-05 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Discovery of Pyridopyrimidinones that Selectively Inhibit the H1047R PI3K alpha Mutant Protein.
J.Med.Chem., 67, 2024
|
|
4CAS
 
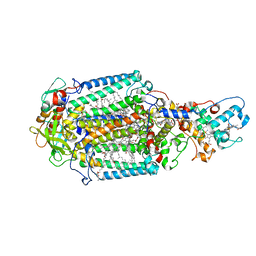 | | Serial femtosecond crystallography structure of a photosynthetic reaction center | | Descriptor: | (2E,6E,10E,14E,18E,22E,26E)-3,7,11,15,19,23,27,31-OCTAMETHYLDOTRIACONTA-2,6,10,14,18,22,26,30-OCTAENYL TRIHYDROGEN DIPHOSPHATE, 15-cis-1,2-dihydroneurosporene, BACTERIOCHLOROPHYLL A, ... | | Authors: | Johansson, L.C, Arnlund, D, Katona, G, White, T.A, Barty, A, DePonte, D.P, Shoeman, R.L, Wickstrand, C, Sharma, A, Williams, G.J, Aquila, A, Bogan, M.J, Caleman, C, Davidsson, J, Doak, R.B, Frank, M, Fromme, R, Galli, L, Grotjohann, I, Hunter, M.S, Kassemeyer, S, Kirian, R.A, Kupitz, C, Liang, M, Lomb, L, Malmerberg, E, Martin, A.V, Messerschmidt, M, Nass, K, Redecke, L, Seibert, M.M, Sjohamn, J, Steinbrener, J, Stellato, F, Wang, D, Wahlgren, W.Y, Weierstall, U, Westenhoff, S, Zatsepin, N.A, Boutet, S, Spence, J.C.H, Schlichting, I, Chapman, H.N, Fromme, P, Neutze, R. | | Deposit date: | 2013-10-09 | | Release date: | 2013-12-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of a photosynthetic reaction centre determined by serial femtosecond crystallography.
Nat Commun, 4, 2013
|
|
1IF0
 
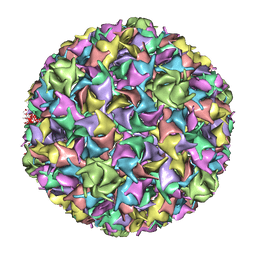 | | PSEUDO-ATOMIC MODEL OF BACTERIOPHAGE HK97 PROCAPSID (PROHEAD II) | | Descriptor: | PROTEIN (MAJOR CAPSID PROTEIN GP5) | | Authors: | Conway, J.F, Wikoff, W.R, Cheng, N, Duda, R.L, Hendrix, R.W, Johnson, J.E, Steven, A.C. | | Deposit date: | 2001-04-11 | | Release date: | 2001-05-02 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Virus maturation involving large subunit rotations and local refolding.
Science, 292, 2001
|
|
1W8O
 
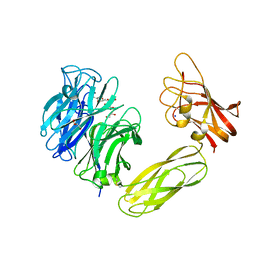 | | Contribution of the Active Site Aspartic Acid to Catalysis in the Bacterial Neuraminidase from Micromonospora viridifaciens | | Descriptor: | BACTERIAL SIALIDASE, CITRIC ACID, GLYCEROL, ... | | Authors: | Newstead, S, Watson, J.N, Dookhun, V, Bennet, A.J, Taylor, G. | | Deposit date: | 2004-09-24 | | Release date: | 2004-09-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Contribution of the Active Site Aspartic Acid to Catalysis in the Bacterial Neuraminidase from Micromonospora Viridifaciens
FEBS Lett., 577, 2004
|
|
1W8N
 
 | | Contribution of the Active Site Aspartic Acid to Catalysis in the Bacterial Neuraminidase from Micromonospora viridifaciens. | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, BACTERIAL SIALIDASE, SODIUM ION, ... | | Authors: | Newstead, S, Watson, J.N, Dookhun, V, Bennet, A.J, Taylor, G. | | Deposit date: | 2004-09-24 | | Release date: | 2004-09-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Contribution of the Active Site Aspartic Acid to Catalysis in the Bacterial Neuraminidase from Micromonospora Viridifaciens
FEBS Lett., 577, 2004
|
|
6DC6
 
 | | Crystal structure of human ubiquitin activating enzyme E1 (Uba1) in complex with ubiquitin | | Descriptor: | MAGNESIUM ION, PYROPHOSPHATE 2-, Ubiquitin, ... | | Authors: | Lv, Z, Yuan, L, Williams, K.M, Atkison, J.H, Olsen, S.K. | | Deposit date: | 2018-05-04 | | Release date: | 2018-10-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.14 Å) | | Cite: | Crystal structure of a human ubiquitin E1-ubiquitin complex reveals conserved functional elements essential for activity.
J. Biol. Chem., 293, 2018
|
|
8GA0
 
 | | CLC-ec1 E202Y at pH 4.5 100mM Cl Turn | | Descriptor: | H(+)/Cl(-) exchange transporter ClcA | | Authors: | Fortea, E, Lee, S, Argyos, Y, Chadda, R, Ciftci, D, Huysmans, G, Robertson, J.L, Boudker, O, Accardi, A. | | Deposit date: | 2023-02-22 | | Release date: | 2024-02-07 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of pH-dependent activation in a CLC transporter.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8GAH
 
 | | CLC-ec1 L25C/A450C/C85A at pH 4.5 100mM Cl Twist | | Descriptor: | CHLORIDE ION, H(+)/Cl(-) exchange transporter ClcA | | Authors: | Fortea, E, Lee, S, Argyos, Y, Chadda, R, Ciftci, D, Huysmans, G, Robertson, J.L, Boudker, O, Accardi, A. | | Deposit date: | 2023-02-22 | | Release date: | 2024-02-07 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of pH-dependent activation in a CLC transporter.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8GA3
 
 | | CLC-ec1 R230C/L249C/C85A at pH 4.5 100mM Cl Turn | | Descriptor: | CHLORIDE ION, H(+)/Cl(-) exchange transporter ClcA | | Authors: | Fortea, E, Lee, S, Argyos, Y, Chadda, R, Ciftci, D, Huysmans, G, Robertson, J.L, Boudker, O, Accardi, A. | | Deposit date: | 2023-02-22 | | Release date: | 2024-02-07 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of pH-dependent activation in a CLC transporter.
Nat.Struct.Mol.Biol., 31, 2024
|
|
2Y44
 
 | | Crystal structure of GARP from Trypanosoma congolense | | Descriptor: | GLUTAMIC ACID/ALANINE-RICH PROTEIN, GLYCEROL, IODIDE ION | | Authors: | Loveless, B.C, Mason, J.W, Sakurai, T, Inoue, N, Razavi, M, Pearson, T.W, Boulanger, M.J. | | Deposit date: | 2011-01-04 | | Release date: | 2011-03-30 | | Last modified: | 2012-11-21 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Characterization and Epitope Mapping of the Glutamic Acid/Alanine-Rich Protein from Trypanosoma Congolense: Defining Assembly on the Parasite Cell Surface.
J.Biol.Chem., 286, 2011
|
|
8AAT
 
 | |
8V8U
 
 | | PI3Ka H1047R co-crystal structure with inhibitor in cryptic pocket near H1047R (compound 12). | | Descriptor: | (2S)-N~1~-{4-methyl-5-[2-(1,1,1-trifluoro-2-methylpropan-2-yl)pyridin-4-yl]-1,3-thiazol-2-yl}pyrrolidine-1,2-dicarboxamide, (3S)-9-[(1R)-1-(2-carboxyanilino)ethyl]-3-cyano-7-methyl-4-oxo-2-(piperidin-1-yl)-3,4-dihydropyrido[1,2-a]pyrimidin-5-ium, Phosphatidylinositol 3-kinase regulatory subunit alpha, ... | | Authors: | Gunn, R.J, Lawson, J.D. | | Deposit date: | 2023-12-06 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Discovery of Pyridopyrimidinones that Selectively Inhibit the H1047R PI3K alpha Mutant Protein.
J.Med.Chem., 67, 2024
|
|
6RIO
 
 | | Imidazole Polyamide-DNA complex NMR structure (5'-CGATGTACATCG-3') | | Descriptor: | 3-[3-[[4-[[4-[[4-[[4-[[(2~{R})-2-azaniumyl-4-[[1-methyl-4-[[1-methyl-4-[[1-methyl-4-[(1-methylimidazol-2-yl)carbonylamino]pyrrol-2-yl]carbonylamino]pyrrol-2-yl]carbonylamino]pyrrol-2-yl]carbonylamino]butanoyl]amino]-1-methyl-imidazol-2-yl]carbonylamino]-1-methyl-pyrrol-2-yl]carbonylamino]-1-methyl-pyrrol-2-yl]carbonylamino]-1-methyl-pyrrol-2-yl]carbonylamino]propanoylamino]propyl-dimethyl-azanium, DNA (5'-(*(DC5)P*GP*AP*TP*GP*TP*AP*CP*AP*TP*CP*(DG3))-3') | | Authors: | Padroni, G, Withers, J.M, Taladriz-Sender, A, Reichenbach, L.F, Parkinson, J.A, Burley, G.A. | | Deposit date: | 2019-04-24 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Sequence-Selective Minor Groove Recognition of a DNA Duplex Containing Synthetic Genetic Components.
J.Am.Chem.Soc., 141, 2019
|
|
2FT1
 
 | | Bacteriophage HK97 Head II | | Descriptor: | major capsid protein | | Authors: | Gan, L, Speir, J.A, Conway, J.F, Lander, G, Cheng, N, Firek, B.A, Hendrix, R.W, Duda, R.L, Liljas, L, Johnson, J.E. | | Deposit date: | 2006-01-23 | | Release date: | 2006-02-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Capsid Conformational Sampling in HK97 Maturation Visualized by X-Ray Crystallography and Cryo-EM.
Structure, 14, 2006
|
|
2C0H
 
 | | X-ray structure of beta-mannanase from blue mussel Mytilus edulis | | Descriptor: | MANNAN ENDO-1,4-BETA-MANNOSIDASE, SULFATE ION | | Authors: | Larsson, A.M, Anderson, L, Xu, B, Munoz, I.G, Uson, I, Janson, J.-C, Stalbrand, H, Stahlberg, J. | | Deposit date: | 2005-09-02 | | Release date: | 2006-02-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Three-Dimensional Crystal Structure and Enzymic Characterization of Beta-Mannanase Man5A from Blue Mussel Mytilus Edulis.
J.Mol.Biol., 357, 2006
|
|
2FS3
 
 | | Bacteriophage HK97 K169Y Head I | | Descriptor: | Major capsid protein | | Authors: | Gan, L, Speir, J.A, Conway, J.F, Lander, G, Cheng, N, Firek, B.A, Hendrix, R.W, Duda, R.L, Liljas, L, Johnson, J.E. | | Deposit date: | 2006-01-20 | | Release date: | 2006-02-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Capsid Conformational Sampling in HK97 Maturation Visualized by X-Ray Crystallography and Cryo-EM.
Structure, 14, 2006
|
|
6CO2
 
 | | Structure of an engineered protein (NUDT16TI) in complex with 53BP1 Tudor domains | | Descriptor: | NUDT16-Tudor-interacting (NUDT16TI), TP53-binding protein 1 | | Authors: | Botuyan, M.V, Thompson, J.R, Cui, G, Mer, G. | | Deposit date: | 2018-03-10 | | Release date: | 2018-06-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Mechanism of 53BP1 activity regulation by RNA-binding TIRR and a designer protein.
Nat. Struct. Mol. Biol., 25, 2018
|
|
