5XV3
 
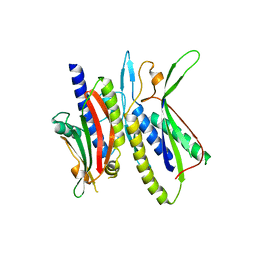 | | Crystal structure of ATG101-ATG13HORMA | | Descriptor: | Autophagy-related protein 101, Autophagy-related protein 13, DI(HYDROXYETHYL)ETHER | | Authors: | Kim, B.-W, Song, H.K. | | Deposit date: | 2017-06-26 | | Release date: | 2018-07-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | The C-terminal region of ATG101 bridges ULK1 and PtdIns3K complex in autophagy initiation.
Autophagy, 14, 2018
|
|
5HZY
 
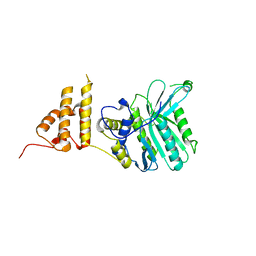 | | Crystal structure of the legionella pneumophila effector protein RavZ - P6322 | | Descriptor: | Uncharacterized protein RavZ | | Authors: | Kwon, D.H, Kim, L, Kim, B.-W, Hong, S.B, Song, H.K. | | Deposit date: | 2016-02-03 | | Release date: | 2016-11-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.548 Å) | | Cite: | The 1:2 complex between RavZ and LC3 reveals a mechanism for deconjugation of LC3 on the phagophore membrane
Autophagy, 13, 2017
|
|
5WT9
 
 | | Complex structure of PD-1 and nivolumab-Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy Chain of Nivolumab, Light Chain of Nivolumab, ... | | Authors: | Tan, S, Zhang, H, Chai, Y, Song, H, Tong, Z, Wang, Q, Qi, J, Wong, G, Zhu, X, Liu, W.J, Gao, S, Wang, Z, Shi, Y, Yang, F, Gao, G.F, Yan, J. | | Deposit date: | 2016-12-10 | | Release date: | 2017-02-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | An unexpected N-terminal loop in PD-1 dominates binding by nivolumab.
Nat Commun, 8, 2017
|
|
5WZ1
 
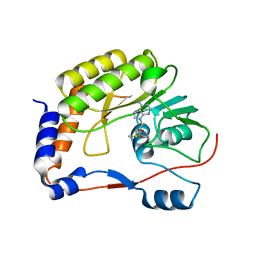 | | Crystal structure of Zika virus NS5 methyltransferase bound to S-adenosyl-L-methionine | | Descriptor: | NS5 methyltransferase, S-ADENOSYLMETHIONINE | | Authors: | Duan, W, Song, H, Qi, J, Shi, Y, Gao, G.F. | | Deposit date: | 2017-01-16 | | Release date: | 2017-03-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.507 Å) | | Cite: | The crystal structure of Zika virus NS5 reveals conserved drug targets.
EMBO J., 36, 2017
|
|
3M6N
 
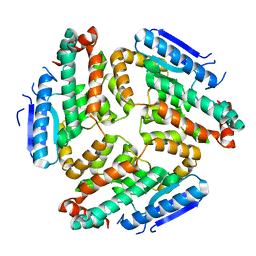 | | Crystal structure of RpfF | | Descriptor: | RpfF protein | | Authors: | Lim, S.C, Cheng, Z, Qamra, R, Song, H. | | Deposit date: | 2010-03-16 | | Release date: | 2010-09-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of the Sensor-Synthase Interaction in Autoinduction of the Quorum Sensing Signal DSF Biosynthesis
Structure, 18, 2010
|
|
5XAD
 
 | | NLIR - LC3B fusion protein | | Descriptor: | Microtubule-associated proteins 1A/1B light chain 3B, Uncharacterised protein | | Authors: | Kwon, D.H, Kim, L, Song, H.K. | | Deposit date: | 2017-03-12 | | Release date: | 2017-07-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | A novel conformation of the LC3-interacting region motif revealed by the structure of a complex between LC3B and RavZ
Biochem. Biophys. Res. Commun., 490, 2017
|
|
3ZYP
 
 | | Cellulose induced protein, Cip1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CIP1, ... | | Authors: | Jacobson, F, Karkehabadi, S, Hansson, H, Goedegebuur, F, Wallace, L, Mitchinson, C, Piens, K, Stals, I, Sandgren, M. | | Deposit date: | 2011-08-24 | | Release date: | 2012-09-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Crystal Structure of the Core Domain of a Cellulose Induced Protein (Cip1) from Hypocrea Jecorina, at 1.5 A Resolution.
Plos One, 8, 2013
|
|
5WZ2
 
 | | Crystal structure of Zika virus NS5 methyltransferase bound to SAM and RNA analogue (m7GpppA) | | Descriptor: | NS5 MTase, P1-7-METHYLGUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE, S-ADENOSYLMETHIONINE | | Authors: | Duan, W, Song, H, Qi, J, Shi, Y, Gao, G.F. | | Deposit date: | 2017-01-16 | | Release date: | 2017-03-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The crystal structure of Zika virus NS5 reveals conserved drug targets.
EMBO J., 36, 2017
|
|
5XGR
 
 | | Structure of the S1 subunit C-terminal domain from bat-derived coronavirus HKU5 spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1 | | Authors: | Xue, H, Qi, J, Song, H, Qihui, W, Shi, Y, Gao, G.F. | | Deposit date: | 2017-04-16 | | Release date: | 2017-05-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the S1 subunit C-terminal domain from bat-derived coronavirus HKU5 spike protein
Virology, 507, 2017
|
|
5XV6
 
 | | Crystal structure of ATG101-ATG13HORMA | | Descriptor: | Autophagy-related protein 101, Autophagy-related protein 13 | | Authors: | Kim, B.-W, Song, H.K. | | Deposit date: | 2017-06-26 | | Release date: | 2018-07-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.455 Å) | | Cite: | The C-terminal region of ATG101 bridges ULK1 and PtdIns3K complex in autophagy initiation.
Autophagy, 14, 2018
|
|
5K66
 
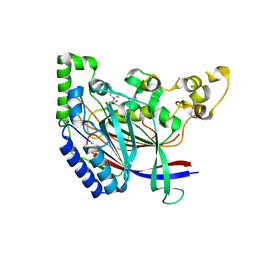 | | Crystal structure of N-terminal amidase with Asn-Glu peptide | | Descriptor: | ASPARAGINE, GLUTAMIC ACID, Nta1p | | Authors: | Kim, M.K, Oh, S.-J, Lee, B.-G, Song, H.K. | | Deposit date: | 2016-05-24 | | Release date: | 2017-01-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Structural basis for dual specificity of yeast N-terminal amidase in the N-end rule pathway.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
3MC0
 
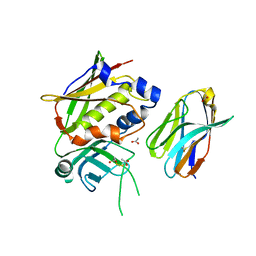 | | Crystal Structure of Staphylococcal Enterotoxin G (SEG) in Complex with a Mouse T-cell Receptor beta Chain | | Descriptor: | ACETATE ION, Enterotoxin SEG, variable beta 8.2 mouse T cell receptor | | Authors: | Fernandez, M.M, Cho, S, Robinson, H, Mariuzza, R.A, Malchiodi, E.L. | | Deposit date: | 2010-03-26 | | Release date: | 2010-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of staphylococcal enterotoxin G (SEG) in complex with a mouse T-cell receptor {beta} chain.
J.Biol.Chem., 286, 2011
|
|
5K62
 
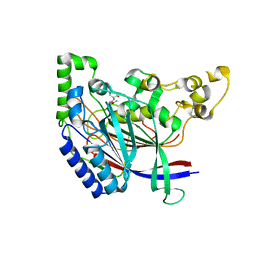 | | Crystal structure of N-terminal amidase C187S | | Descriptor: | ASPARAGINE, Nta1p, VALINE | | Authors: | Kim, M.K, Oh, S.-J, Lee, B.-G, Song, H.K. | | Deposit date: | 2016-05-24 | | Release date: | 2017-01-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | Structural basis for dual specificity of yeast N-terminal amidase in the N-end rule pathway.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
5YSI
 
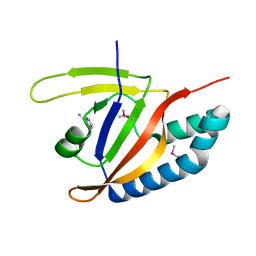 | | SdeA mART-C domain EE/AA NCA complex | | Descriptor: | NICOTINAMIDE, Ubiquitinating/deubiquitinating enzyme SdeA | | Authors: | Kim, L, Kwon, D.H, Song, H.K. | | Deposit date: | 2017-11-14 | | Release date: | 2018-08-29 | | Method: | X-RAY DIFFRACTION (1.546 Å) | | Cite: | Structural and Biochemical Study of the Mono-ADP-Ribosyltransferase Domain of SdeA, a Ubiquitylating/Deubiquitylating Enzyme from Legionella pneumophila
J. Mol. Biol., 430, 2018
|
|
7BZN
 
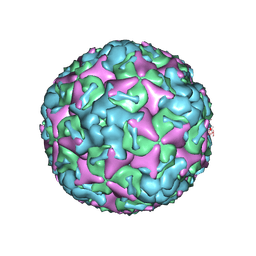 | | Cryo-EM structure of mature Coxsackievirus A10 at pH 7.4 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Cui, Y, Peng, R, Song, H, Tong, Z, Gao, G.F, Qi, J. | | Deposit date: | 2020-04-28 | | Release date: | 2020-07-22 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular basis of Coxsackievirus A10 entry using the two-in-one attachment and uncoating receptor KRM1.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7BZO
 
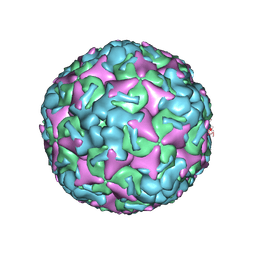 | | Cryo-EM structure of mature Coxsackievirus A10 at pH 5.5 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Cui, Y, Peng, R, Song, H, Tong, Z, Gao, G.F, Qi, J. | | Deposit date: | 2020-04-28 | | Release date: | 2020-07-22 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular basis of Coxsackievirus A10 entry using the two-in-one attachment and uncoating receptor KRM1.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
3NIL
 
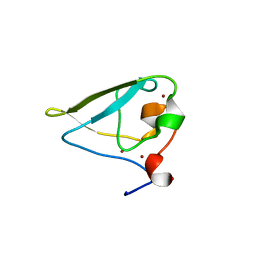 | | The structure of UBR box (RDAA) | | Descriptor: | ACETATE ION, E3 ubiquitin-protein ligase UBR1, Peptide RDAA, ... | | Authors: | Choi, W.S, Jeong, B.-C, Lee, M.-R, Song, H.K. | | Deposit date: | 2010-06-16 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for the recognition of N-end rule substrates by the UBR box of ubiquitin ligases
Nat.Struct.Mol.Biol., 17, 2010
|
|
3NIK
 
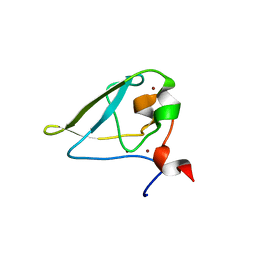 | | The structure of UBR box (REAA) | | Descriptor: | E3 ubiquitin-protein ligase UBR1, Peptide REAA, ZINC ION | | Authors: | Choi, W.S, Jeong, B.-C, Lee, M.-R, Song, H.K. | | Deposit date: | 2010-06-16 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for the recognition of N-end rule substrates by the UBR box of ubiquitin ligases
Nat.Struct.Mol.Biol., 17, 2010
|
|
3NIS
 
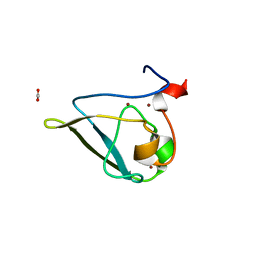 | | The structure of UBR box (native2) | | Descriptor: | ACETATE ION, E3 ubiquitin-protein ligase UBR1, ZINC ION | | Authors: | Choi, W.S, Jeong, B.-C, Lee, M.-R, Song, H.K. | | Deposit date: | 2010-06-16 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural basis for the recognition of N-end rule substrates by the UBR box of ubiquitin ligases
Nat.Struct.Mol.Biol., 17, 2010
|
|
5K5U
 
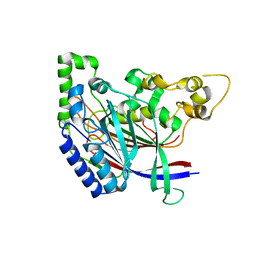 | |
5K63
 
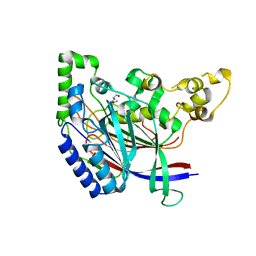 | | Crystal structure of N-terminal amidase C187S | | Descriptor: | ASPARAGINE, GLYCINE, Nta1p | | Authors: | Kim, M.K, Oh, S.-J, Lee, B.-G, Song, H.K. | | Deposit date: | 2016-05-24 | | Release date: | 2017-01-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for dual specificity of yeast N-terminal amidase in the N-end rule pathway.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
7BV7
 
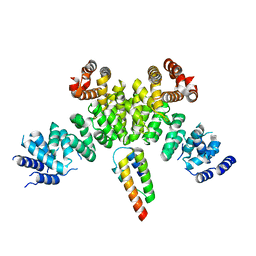 | | INTS3 complexed with INTS6 | | Descriptor: | Integrator complex subunit 3, Integrator complex subunit 6 | | Authors: | Jia, Y, Bharath, S.R, Song, H. | | Deposit date: | 2020-04-09 | | Release date: | 2021-07-14 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the INTS3/INTS6 complex reveals the functional importance of INTS3 dimerization in DSB repair.
Cell Discov, 7, 2021
|
|
3NIM
 
 | | The structure of UBR box (RRAA) | | Descriptor: | E3 ubiquitin-protein ligase UBR1, Peptide RRAA, ZINC ION | | Authors: | Choi, W.S, Jeong, B.-C, Lee, M.-R, Song, H.K. | | Deposit date: | 2010-06-16 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the recognition of N-end rule substrates by the UBR box of ubiquitin ligases
Nat.Struct.Mol.Biol., 17, 2010
|
|
2HUI
 
 | | Crystal structure of Aedes aegypti alanine glyoxylate aminotransferase in complex with glyoxylic acid | | Descriptor: | Alanine glyoxylate aminotransferase, GLYOXYLIC ACID | | Authors: | Han, Q, Robinson, H, Gao, Y.G, Vogelaar, N, Wilson, S.R, Rizzi, M, Li, J. | | Deposit date: | 2006-07-26 | | Release date: | 2006-09-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structures of Aedes aegypti Alanine Glyoxylate Aminotransferase.
J.Biol.Chem., 281, 2006
|
|
2HUU
 
 | | Crystal structure of Aedes aegypti alanine glyoxylate aminotransferase in complex with alanine | | Descriptor: | 1-BUTANOL, ALANINE, Alanine glyoxylate aminotransferase | | Authors: | Han, Q, Robinson, H, Gao, Y.G, Vogelaar, N, Wilson, S.R, Rizzi, M, Li, J. | | Deposit date: | 2006-07-27 | | Release date: | 2006-09-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of Aedes aegypti Alanine Glyoxylate Aminotransferase.
J.Biol.Chem., 281, 2006
|
|
