7Y70
 
 | | RLGSGG-AtPRT6 UBR box (P4332) | | 分子名称: | E3 ubiquitin-protein ligase PRT6, ZINC ION | | 著者 | Kim, L, Song, H.K. | | 登録日 | 2022-06-21 | | 公開日 | 2023-07-05 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.801 Å) | | 主引用文献 | Structural analyses of plant PRT6-UBR box for Cys-Arg/N-degron pathway and insights into the plant submergence resistance
To Be Published
|
|
7XWD
 
 | | Apo-AtPRT6 UBR box | | 分子名称: | E3 ubiquitin-protein ligase PRT6, ZINC ION | | 著者 | Kim, L, Song, H.K. | | 登録日 | 2022-05-26 | | 公開日 | 2023-05-31 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.396 Å) | | 主引用文献 | Structural analyses of plant PRT6-UBR box for Cys-Arg/N-degron pathway and insights into the plant submergence resistance
To Be Published
|
|
6XJO
 
 | |
7Y6Z
 
 | | RLGSGG-AtPRT6 UBR box (I222) | | 分子名称: | E3 ubiquitin-protein ligase PRT6, ZINC ION | | 著者 | Kim, L, Song, H.K. | | 登録日 | 2022-06-21 | | 公開日 | 2023-07-05 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.598 Å) | | 主引用文献 | Structural analyses of plant PRT6-UBR box for Cys-Arg/N-degron pathway and insights into the plant submergence resistance
To Be Published
|
|
5XV4
 
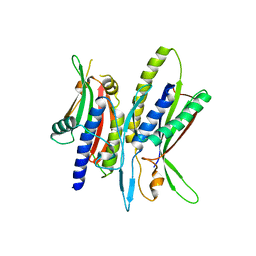 | | Crystal structure of ATG101-ATG13HORMA | | 分子名称: | Autophagy-related protein 101, Autophagy-related protein 13 | | 著者 | Kim, B.-W, Song, H.K. | | 登録日 | 2017-06-26 | | 公開日 | 2018-07-04 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.95 Å) | | 主引用文献 | The C-terminal region of ATG101 bridges ULK1 and PtdIns3K complex in autophagy initiation.
Autophagy, 14, 2018
|
|
6ICO
 
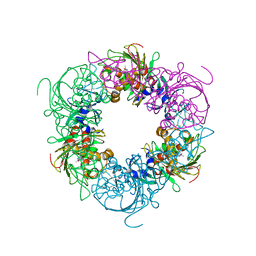 | | Pseudomonas putida CBB5 NdmA with theophylline | | 分子名称: | COBALT (II) ION, FE2/S2 (INORGANIC) CLUSTER, Methylxanthine N1-demethylase NdmA, ... | | 著者 | Kim, J.H, Kim, B.H, Kang, S.Y, Song, H.K. | | 登録日 | 2018-09-06 | | 公開日 | 2019-09-04 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structural and Mechanistic Insights into Caffeine Degradation by the Bacterial N-Demethylase Complex.
J.Mol.Biol., 431, 2019
|
|
6ICM
 
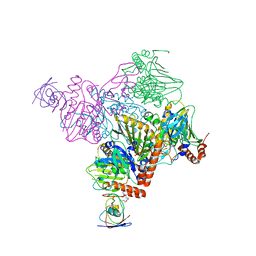 | | Pseudomonas putida CBB5 NdmA with ferredoxin domain of NdmD | | 分子名称: | FE (III) ION, FE2/S2 (INORGANIC) CLUSTER, Methylxanthine N1-demethylase NdmA, ... | | 著者 | Kim, J.H, Kim, B.H, Kang, S.Y, Song, H.K. | | 登録日 | 2018-09-06 | | 公開日 | 2019-09-04 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.961 Å) | | 主引用文献 | Structural and Mechanistic Insights into Caffeine Degradation by the Bacterial N-Demethylase Complex.
J.Mol.Biol., 431, 2019
|
|
6ICK
 
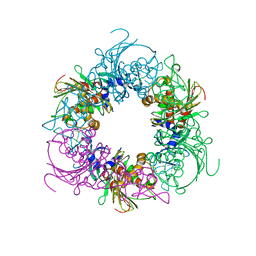 | | Pseudomonas putida CBB5 NdmA | | 分子名称: | FE (III) ION, FE2/S2 (INORGANIC) CLUSTER, Methylxanthine N1-demethylase NdmA | | 著者 | Kim, J.H, Kim, B.H, Kang, S.Y, Song, H.K. | | 登録日 | 2018-09-06 | | 公開日 | 2019-09-04 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.952 Å) | | 主引用文献 | Structural and Mechanistic Insights into Caffeine Degradation by the Bacterial N-Demethylase Complex.
J.Mol.Biol., 431, 2019
|
|
8PVA
 
 | | Structure of bacterial ribosome determined by cryoEM at 100 keV | | 分子名称: | 16S rRNA, 23S rRNA, 50S ribosomal protein L14, ... | | 著者 | McMullan, G, Naydenova, K, Mihaylov, D, Peet, M.J, Wilson, H, Yamashita, K, Dickerson, J.L, Chen, S, Cannone, G, Lee, Y, Hutchings, K.A, Gittins, O, Sobhy, M, Wells, T, El-Gomati, M.M, Dalby, J, Meffert, M, Schulze-Briese, C, Henderson, R, Russo, C.J. | | 登録日 | 2023-07-17 | | 公開日 | 2023-11-29 | | 最終更新日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | Structure determination by cryoEM at 100 keV.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8PVB
 
 | | Structure of GABAAR determined by cryoEM at 100 keV | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, DECANE, ... | | 著者 | McMullan, G, Naydenova, K, Mihaylov, D, Peet, M.J, Wilson, H, Yamashita, K, Dickerson, J.L, Chen, S, Cannone, G, Lee, Y, Hutchings, K.A, Gittins, O, Sobhy, M, Wells, T, El-Gomati, M.M, Dalby, J, Meffert, M, Schulze-Briese, C, Henderson, R, Russo, C.J. | | 登録日 | 2023-07-17 | | 公開日 | 2023-11-29 | | 最終更新日 | 2024-01-31 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structure determination by cryoEM at 100 keV.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7YRB
 
 | | UBR box of human UBR6 | | 分子名称: | F-box protein 11, isoform CRA_f, SULFATE ION, ... | | 著者 | Kim, B, Song, H.K. | | 登録日 | 2022-08-09 | | 公開日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.51 Å) | | 主引用文献 | Crystal structure of UBR box from human UBR6
To Be Published
|
|
7Y6W
 
 | | RRGSGG-AtPRT6 UBR box (I222) | | 分子名称: | E3 ubiquitin-protein ligase PRT6, ZINC ION | | 著者 | Kim, L, Song, H.K. | | 登録日 | 2022-06-21 | | 公開日 | 2023-07-05 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structural analyses of plant PRT6-UBR box for Cys-Arg/N-degron pathway and insights into the plant submergence resistance
To Be Published
|
|
7X0F
 
 | |
7X17
 
 | | Structure of Pseudomonas NRPS protein, AmbB-TC bound to Ppant-L-Ala | | 分子名称: | AMB antimetabolite synthase AmbB, S-[2-[3-[[(2S)-3,3-dimethyl-2-oxidanyl-4-phosphonooxy-butanoyl]amino]propanoylamino]ethyl] (2R)-2-azanylpropanethioate | | 著者 | ChuYuanKee, M, Bharath, S.R, Song, H. | | 登録日 | 2022-02-23 | | 公開日 | 2022-07-13 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural insights into the substrate-bound condensation domains of non-ribosomal peptide synthetase AmbB.
Sci Rep, 12, 2022
|
|
6YYE
 
 | | TREM2 extracellular domain (19-131) in complex with single-chain variable fragment (scFv-2) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, TREM2 Single chain variable 2, Triggering receptor expressed on myeloid cells 2 | | 著者 | Szykowska, A, Preger, C, Krojer, T, Mukhopadhyay, S.M.M, McKinley, G, Graslund, S, Wigren, E, Persson, H, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Di Daniel, E, Burgess-Brown, N, Bullock, A. | | 登録日 | 2020-05-04 | | 公開日 | 2021-02-17 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (3.36 Å) | | 主引用文献 | Selection and structural characterization of anti-TREM2 scFvs that reduce levels of shed ectodomain.
Structure, 29, 2021
|
|
4TYX
 
 | | Structure of aquoferric sperm whale myoglobin L29H/F33Y/F43H/S92A mutant | | 分子名称: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Bhagi-Damodaran, A, Petrik, I.D, Robinson, H, Lu, Y. | | 登録日 | 2014-07-09 | | 公開日 | 2014-08-13 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.64 Å) | | 主引用文献 | Systematic tuning of heme redox potentials and its effects on O2 reduction rates in a designed oxidase in myoglobin.
J.Am.Chem.Soc., 136, 2014
|
|
5XJD
 
 | | TEAD in complex with fragment | | 分子名称: | (2S)-2-phenyl-2-pyrrol-1-yl-ethanoic acid, Transcriptional enhancer factor TEF-3 | | 著者 | Kaan, H.Y.K, Sim, A.Y.L, Tan, S.K.J, Verma, C, Song, H. | | 登録日 | 2017-05-01 | | 公開日 | 2018-01-24 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.22 Å) | | 主引用文献 | Targeting YAP/TAZ-TEAD protein-protein interactions using fragment-based and computational modeling approaches.
PLoS ONE, 12, 2017
|
|
5XAC
 
 | | CLIR - LC3B | | 分子名称: | Microtubule-associated proteins 1A/1B light chain 3B | | 著者 | Kwon, D.H, Kim, L, Song, H.K. | | 登録日 | 2017-03-12 | | 公開日 | 2017-07-12 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.701 Å) | | 主引用文献 | A novel conformation of the LC3-interacting region motif revealed by the structure of a complex between LC3B and RavZ
Biochem. Biophys. Res. Commun., 490, 2017
|
|
5XAE
 
 | | mutNLIR_LC3B | | 分子名称: | Microtubule-associated proteins 1A/1B light chain 3B | | 著者 | Kwon, D.H, Kim, L, Song, H.K. | | 登録日 | 2017-03-12 | | 公開日 | 2017-07-12 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.996 Å) | | 主引用文献 | A novel conformation of the LC3-interacting region motif revealed by the structure of a complex between LC3B and RavZ
Biochem. Biophys. Res. Commun., 490, 2017
|
|
5XAD
 
 | | NLIR - LC3B fusion protein | | 分子名称: | Microtubule-associated proteins 1A/1B light chain 3B, Uncharacterised protein | | 著者 | Kwon, D.H, Kim, L, Song, H.K. | | 登録日 | 2017-03-12 | | 公開日 | 2017-07-12 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.88 Å) | | 主引用文献 | A novel conformation of the LC3-interacting region motif revealed by the structure of a complex between LC3B and RavZ
Biochem. Biophys. Res. Commun., 490, 2017
|
|
6SJ4
 
 | | Amidohydrolase, AHS with substrate analog | | 分子名称: | 1,2-ETHANEDIOL, 3-(3-hydroxyphenyl)carbonyloxybenzoic acid, Amidohydrolase, ... | | 著者 | Naismith, J.H, Song, H. | | 登録日 | 2019-08-12 | | 公開日 | 2020-01-15 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.81 Å) | | 主引用文献 | The Biosynthesis of the Benzoxazole in Nataxazole Proceeds via an Unstable Ester and has Synthetic Utility.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
5WT9
 
 | | Complex structure of PD-1 and nivolumab-Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy Chain of Nivolumab, Light Chain of Nivolumab, ... | | 著者 | Tan, S, Zhang, H, Chai, Y, Song, H, Tong, Z, Wang, Q, Qi, J, Wong, G, Zhu, X, Liu, W.J, Gao, S, Wang, Z, Shi, Y, Yang, F, Gao, G.F, Yan, J. | | 登録日 | 2016-12-10 | | 公開日 | 2017-02-15 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.401 Å) | | 主引用文献 | An unexpected N-terminal loop in PD-1 dominates binding by nivolumab.
Nat Commun, 8, 2017
|
|
5YPC
 
 | | p62/SQSTM1 ZZ domain with Phe-peptide | | 分子名称: | 78 kDa glucose-regulated protein,Sequestosome-1, ZINC ION | | 著者 | Kwon, D.H, Kim, L, Song, H.K. | | 登録日 | 2017-11-01 | | 公開日 | 2018-08-29 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.962 Å) | | 主引用文献 | Insights into degradation mechanism of N-end rule substrates by p62/SQSTM1 autophagy adapter.
Nat Commun, 9, 2018
|
|
8PVC
 
 | | Structure of mouse heavy-chain apoferritin determined by cryoEM at 100 keV | | 分子名称: | FE (III) ION, Ferritin heavy chain, ZINC ION | | 著者 | McMullan, G, Naydenova, K, Mihaylov, D, Peet, M.J, Wilson, H, Yamashita, K, Dickerson, J.L, Chen, S, Cannone, G, Lee, Y, Hutchings, K.A, Gittins, O, Sobhy, M, Wells, T, El-Gomati, M.M, Dalby, J, Meffert, M, Schulze-Briese, C, Henderson, R, Russo, C.J. | | 登録日 | 2023-07-17 | | 公開日 | 2023-11-29 | | 最終更新日 | 2023-12-06 | | 実験手法 | ELECTRON MICROSCOPY (2.6 Å) | | 主引用文献 | Structure determination by cryoEM at 100 keV.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5YPB
 
 | | p62/SQSTM1 ZZ domain with His-peptide | | 分子名称: | 78 kDa glucose-regulated protein,Sequestosome-1, ZINC ION | | 著者 | Kwon, D.H, Kim, L, Song, H.K. | | 登録日 | 2017-11-01 | | 公開日 | 2018-08-29 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Insights into degradation mechanism of N-end rule substrates by p62/SQSTM1 autophagy adapter.
Nat Commun, 9, 2018
|
|
