3C7A
 
 | | A structural basis for substrate and stereo selectivity in octopine dehydrogenase (ODH-NADH) | | Descriptor: | 1,2-ETHANEDIOL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Octopine dehydrogenase | | Authors: | Smits, S.H.J, Mueller, A, Schmitt, L, Grieshaber, M.K. | | Deposit date: | 2008-02-07 | | Release date: | 2008-07-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A structural basis for substrate selectivity and stereoselectivity in octopine dehydrogenase from Pecten maximus.
J.Mol.Biol., 381, 2008
|
|
3C7D
 
 | | A structural basis for substrate and stereo selectivity in octopine dehydrogenase (ODH-NADH-Pyruvate) | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Octopine dehydrogenase, PYRUVIC ACID | | Authors: | Smits, S.H.J, Mueller, A, Schmitt, L, Grieshaber, M.K. | | Deposit date: | 2008-02-07 | | Release date: | 2008-07-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A structural basis for substrate selectivity and stereoselectivity in octopine dehydrogenase from Pecten maximus.
J.Mol.Biol., 381, 2008
|
|
3C7C
 
 | | A structural basis for substrate and stereo selectivity in octopine dehydrogenase (ODH-NADH-L-Arginine) | | Descriptor: | ARGININE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Octopine dehydrogenase | | Authors: | Smits, S.H.J, Mueller, A, Schmitt, L, Grieshaber, M.K. | | Deposit date: | 2008-02-07 | | Release date: | 2008-07-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A structural basis for substrate selectivity and stereoselectivity in octopine dehydrogenase from Pecten maximus.
J.Mol.Biol., 381, 2008
|
|
3IQD
 
 | | Structure of Octopine-dehydrogenase in complex with NADH and Agmatine | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, AGMATINE, Octopine dehydrogenase | | Authors: | Smits, S.H.J, Meyer, T, Mueller, A, Willbold, D, Grieshaber, M.K, Schmitt, L. | | Deposit date: | 2009-08-20 | | Release date: | 2010-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Insights into the mechanism of ligand binding to octopine dehydrogenase from Pecten maximus by NMR and crystallography
Plos One, 5, 2010
|
|
3CHG
 
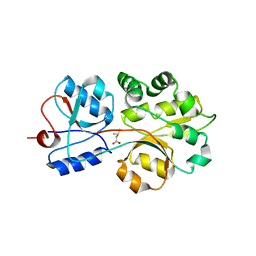 | | The compatible solute-binding protein OpuAC from Bacillus subtilis in complex with DMSA | | Descriptor: | (dimethyl-lambda~4~-sulfanyl)acetic acid, Glycine betaine-binding protein | | Authors: | Smits, S.H.J, Hoing, M, Lecher, J, Jebbar, M, Schmitt, L, Bremer, E. | | Deposit date: | 2008-03-09 | | Release date: | 2008-08-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Compatible-Solute-Binding Protein OpuAC from Bacillus subtilis: Ligand Binding, Site-Directed Mutagenesis, and Crystallographic Studies
J.Bacteriol., 190, 2008
|
|
8A2C
 
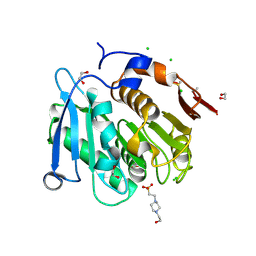 | | The crystal structure of the S178A mutant of PET40, a PETase enzyme from an unclassified Amycolatopsis | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Costanzi, E, Applegate, V, Port, A, Smits, S.H.J. | | Deposit date: | 2022-06-03 | | Release date: | 2023-06-14 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The metagenome-derived esterase PET40 is highly promiscuous and hydrolyses polyethylene terephthalate (PET).
Febs J., 291, 2024
|
|
3R6U
 
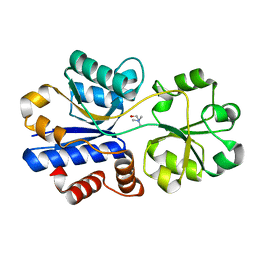 | | Crystal structure of choline binding protein OpuBC from Bacillus subtilis | | Descriptor: | CHOLINE ION, Choline-binding protein | | Authors: | Pittelkow, M, Tschapek, B, Smits, S.H.J, Schmitt, L, Bremer, E. | | Deposit date: | 2011-03-22 | | Release date: | 2011-06-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | The Crystal Structure of the Substrate-Binding Protein OpuBC from Bacillus subtilis in Complex with Choline.
J.Mol.Biol., 411, 2011
|
|
1I1G
 
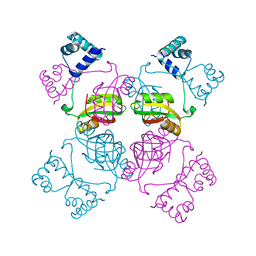 | | CRYSTAL STRUCTURE OF THE LRP-LIKE TRANSCRIPTIONAL REGULATOR FROM THE ARCHAEON PYROCOCCUS FURIOSUS | | Descriptor: | TRANSCRIPTIONAL REGULATOR LRPA | | Authors: | Leonard, P.M, Smits, S.H.J, Sedelnikova, S.E, Brinkman, A.B, de Vos, W.M, van der Oost, J, Rice, D.W, Rafferty, J.B. | | Deposit date: | 2001-02-01 | | Release date: | 2002-02-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the Lrp-like transcriptional regulator from the archaeon Pyrococcus furiosus.
EMBO J., 20, 2001
|
|
3HCQ
 
 | | Structural analysis of the choline binding protein ChoX in a semi-closed and ligand-free conformation | | Descriptor: | Putative choline ABC transporter, periplasmic solute-binding component | | Authors: | Oswald, C, Smits, S.H.J, Hoeing, M, Bremer, E, Schmitt, L. | | Deposit date: | 2009-05-06 | | Release date: | 2009-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structural analysis of the choline-binding protein ChoX in a semi-closed and ligand-free conformation.
Biol.Chem., 390, 2009
|
|
8B4U
 
 | | The crystal structure of PET46, a PETase enzyme from Candidatus bathyarchaeota | | Descriptor: | 1,2-ETHANEDIOL, Alpha/beta hydrolase, CHLORIDE ION, ... | | Authors: | Costanzi, E, Applegate, V, Schumacher, J, Smits, S.H.J. | | Deposit date: | 2022-09-21 | | Release date: | 2023-08-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | An archaeal lid-containing feruloyl esterase degrades polyethylene terephthalate.
Commun Chem, 6, 2023
|
|
3FXB
 
 | | Crystal structure of the ectoine-binding protein UehA | | Descriptor: | (4S)-2-METHYL-1,4,5,6-TETRAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, TRAP dicarboxylate transporter, DctP subunit | | Authors: | Lecher, J, Pittelkow, M, Bursy, J, Smits, S.H.J, Schmitt, L, Bremer, E. | | Deposit date: | 2009-01-20 | | Release date: | 2009-05-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The crystal structure of UehA in complex with ectoine-A comparison with other TRAP-T binding proteins.
J.Mol.Biol., 389, 2009
|
|
6Z68
 
 | | A novel metagenomic alpha/beta-fold esterase | | Descriptor: | Acetyl esterase/lipase, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Bollinger, A, Thies, S, Hoeppner, A, Kobus, S, Jaeger, K.-E, Smits, S.H.J. | | Deposit date: | 2020-05-28 | | Release date: | 2020-12-30 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structures of a novel family IV esterase in free and substrate-bound form.
Febs J., 288, 2021
|
|
6Z69
 
 | | A novel metagenomic alpha/beta-fold esterase | | Descriptor: | 7-hydroxy-4-methyl-2H-chromen-2-one, Acetyl esterase/lipase, MAGNESIUM ION, ... | | Authors: | Bollinger, A, Thies, S, Hoeppner, A, Kobus, S, Jaeger, K.-E, Smits, S.H.J. | | Deposit date: | 2020-05-28 | | Release date: | 2020-12-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystal structures of a novel family IV esterase in free and substrate-bound form.
Febs J., 288, 2021
|
|
5BY5
 
 | | High resolution structure of the ectoine synthase from the cold-adapted marine bacterium Sphingopyxis alaskensis | | Descriptor: | L-ectoine synthase, S-1,2-PROPANEDIOL | | Authors: | Widderich, N, Kobus, S, Hoeppner, A, Bremer, E, Smits, S.H.J. | | Deposit date: | 2015-06-10 | | Release date: | 2016-04-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Biochemistry and Crystal Structure of Ectoine Synthase: A Metal-Containing Member of the Cupin Superfamily.
Plos One, 11, 2016
|
|
5DCM
 
 | | Structure of a lantibiotic response regulator: C-terminal domain of the nisin resistance regulator NsrR | | Descriptor: | PhoB family transcriptional regulator | | Authors: | Khosa, S, Kleinschrodt, D, Hoeppner, A, Smits, S.H.J. | | Deposit date: | 2015-08-24 | | Release date: | 2016-07-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the Response Regulator NsrR from Streptococcus agalactiae, Which Is Involved in Lantibiotic Resistance.
Plos One, 11, 2016
|
|
5BXX
 
 | | Crystal structure of the ectoine synthase from the cold-adapted marine bacterium Sphingopyxis alaskensis | | Descriptor: | L-ectoine synthase | | Authors: | Widderich, N, Kobus, S, Hoeppner, A, Bremer, E, Smits, S.H.J. | | Deposit date: | 2015-06-09 | | Release date: | 2016-04-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Biochemistry and Crystal Structure of Ectoine Synthase: A Metal-Containing Member of the Cupin Superfamily.
Plos One, 11, 2016
|
|
8B6E
 
 | | crystal structure of the DNA-binding short chromatophore-targeted protein sCTP-23166 from Paulinella chromatophora | | Descriptor: | 1,2-ETHANEDIOL, SODIUM ION, sCTP-23166 | | Authors: | Macorano, L, Applegate, V, Hoeppner, A, Smits, S.H.J, Nowack, E.C.M. | | Deposit date: | 2022-09-27 | | Release date: | 2023-07-12 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | DNA-binding and protein structure of nuclear factors likely acting in genetic information processing in the Paulinella chromatophore.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
3B5J
 
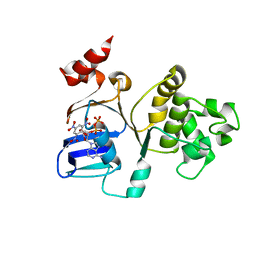 | | Crystal Structures of the S504A Mutant of an Isolated ABC-ATPase in Complex with TNP-ADP | | Descriptor: | 2',3'-O-[(1R,6R)-2,4,6-trinitrocyclohexa-2,4-diene-1,1-diyl]adenosine 5'-(trihydrogen diphosphate), Alpha-hemolysin translocation ATP-binding protein hlyB | | Authors: | Oswald, C, Jenewein, S, Smits, S.H.J, Holland, I.B, Schmitt, L. | | Deposit date: | 2007-10-26 | | Release date: | 2008-01-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Water-mediated protein-fluorophore interactions modulate the affinity of an ABC-ATPase/TNP-ADP complex
J.Struct.Biol., 162, 2008
|
|
2REG
 
 | | ABC-transporter choline binding protein in complex with choline | | Descriptor: | CHOLINE ION, PUTATIVE GLYCINE BETAINE-BINDING ABC TRANSPORTER PROTEIN | | Authors: | Oswald, C, Schimtt, L, Smits, S.H.J, Hoeing, M, Sohn-Boeser, L, Bremer, E. | | Deposit date: | 2007-09-26 | | Release date: | 2008-09-16 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of the choline/acetylcholine substrate-binding protein ChoX from Sinorhizobium meliloti in the liganded and unliganded-closed states.
J.Biol.Chem., 283, 2008
|
|
2REJ
 
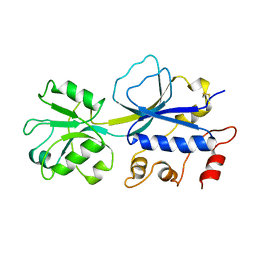 | | ABC-transporter choline binding protein in unliganded semi-closed conformation | | Descriptor: | PUTATIVE GLYCINE BETAINE ABC TRANSPORTER PROTEIN | | Authors: | Oswald, C, Smits, S.H.J, Hoeing, M, Sohn-Boeser, L, Le Rudulier, D, Schmitt, L, Bremer, E. | | Deposit date: | 2007-09-26 | | Release date: | 2008-09-16 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of the choline/acetylcholine substrate-binding protein ChoX from Sinorhizobium meliloti in the liganded and unliganded-closed states.
J.Biol.Chem., 283, 2008
|
|
2RF1
 
 | | Crystal structure of ChoX in an unliganded closed conformation | | Descriptor: | PUTATIVE GLYCINE BETAINE-BINDING ABC TRANSPORTER PROTEIN | | Authors: | Oswald, C, Smits, S.H.J, Hoeing, M, Sohn-Boeser, L, Le Rudulier, D, Schmitt, L, Bremer, E. | | Deposit date: | 2007-09-27 | | Release date: | 2008-09-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of the choline/acetylcholine substrate-binding protein ChoX from Sinorhizobium meliloti in the liganded and unliganded-closed states.
J.Biol.Chem., 283, 2008
|
|
2RIN
 
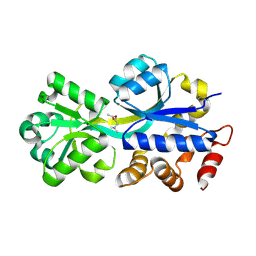 | | ABC-transporter choline binding protein in complex with acetylcholine | | Descriptor: | ACETYLCHOLINE, PUTATIVE GLYCINE BETAINE-BINDING ABC TRANSPORTER PROTEIN | | Authors: | Oswald, C, Smits, S.H.J, Hoeing, M, Sohn-Boeser, L, Le Rudulier, D, Schmitt, L, Bremer, E. | | Deposit date: | 2007-10-12 | | Release date: | 2008-09-30 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of the choline/acetylcholine substrate-binding protein ChoX from Sinorhizobium meliloti in the liganded and unliganded-closed states.
J.Biol.Chem., 283, 2008
|
|
6SCD
 
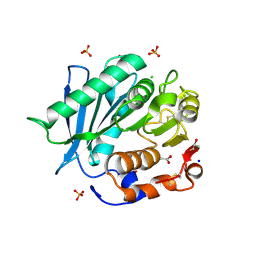 | | Polyester hydrolase PE-H Y250S mutant of Pseudomonas aestusnigri | | Descriptor: | ACETATE ION, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Bollinger, A, Thies, S, Kobus, S, Hoeppner, A, Smits, S.H.J, Jaeger, K.-E. | | Deposit date: | 2019-07-24 | | Release date: | 2020-02-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | A Novel Polyester Hydrolase From the Marine BacteriumPseudomonas aestusnigri -Structural and Functional Insights.
Front Microbiol, 11, 2020
|
|
6SBN
 
 | | Polyester hydrolase PE-H of Pseudomonas aestusnigri | | Descriptor: | ACETATE ION, SODIUM ION, polyester hydrolase | | Authors: | Bollinger, A, Thies, S, Kobus, S, Hoeppner, A, Smits, S.H.J, Jaeger, K.-E. | | Deposit date: | 2019-07-22 | | Release date: | 2020-02-26 | | Last modified: | 2020-03-11 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | A Novel Polyester Hydrolase From the Marine BacteriumPseudomonas aestusnigri -Structural and Functional Insights.
Front Microbiol, 11, 2020
|
|
6SL8
 
 | | Diaminobutyrate acetyltransferase EctA from Paenibacillus lautus in complex with its substrate L-2,4-diaminobutyric acid (DAB) | | Descriptor: | 2,4-DIAMINOBUTYRIC ACID, GLYCEROL, L-2,4-diaminobutyric acid acetyltransferase, ... | | Authors: | Richter, A.A, Kobus, S, Czech, L, Hoeppner, A, Bremer, E, Smits, S.H.J. | | Deposit date: | 2019-08-19 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | The architecture of the diaminobutyrate acetyltransferase active site provides mechanistic insight into the biosynthesis of the chemical chaperone ectoine.
J.Biol.Chem., 295, 2020
|
|
