7LVZ
 
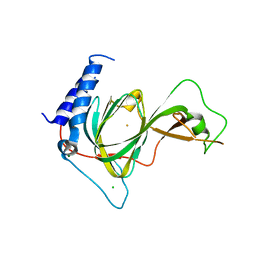 | | Crystal structure of ADO | | Descriptor: | 2-aminoethanethiol dioxygenase, CHLORIDE ION, FE (II) ION, ... | | Authors: | Bingman, C.A, Fernandez, R.L, Smith, R.W, Fox, B.G, Brunold, T.C. | | Deposit date: | 2021-02-26 | | Release date: | 2022-01-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | The Crystal Structure of Cysteamine Dioxygenase Reveals the Origin of the Large Substrate Scope of This Vital Mammalian Enzyme.
Biochemistry, 60, 2021
|
|
6AWL
 
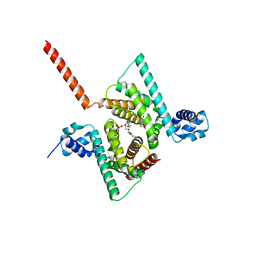 | | Crystal structure of human Coq9 | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI-PALMITOYL-3-SN-PHOSPHATIDYLETHANOLAMINE, Ubiquinone biosynthesis protein COQ9, ... | | Authors: | Bingman, C.A, Lohman, D.C, Smith, R.W, Pagliarini, D.J, Mitochondrial Protein Partnership (MPP) | | Deposit date: | 2017-09-05 | | Release date: | 2019-02-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An Isoprene Lipid-Binding Protein Promotes Eukaryotic Coenzyme Q Biosynthesis.
Mol. Cell, 73, 2019
|
|
8DK0
 
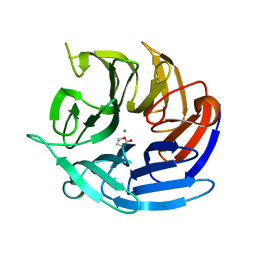 | | Crystal structure of RPA3624, a beta-propeller lactonase from Rhodopseudomonas palustris, with active-site bound (S)gamma-valerolactone | | Descriptor: | CALCIUM ION, Gluconolactonase, SODIUM ION, ... | | Authors: | Bingman, C.A, Hall, B.W, Smith, R.W, Fox, B.G, Donohue, T.J. | | Deposit date: | 2022-07-01 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A broad specificity beta-propeller enzyme from Rhodopseudomonas palustris that hydrolyzes many lactones including gamma-valerolactone.
J.Biol.Chem., 299, 2022
|
|
6UI3
 
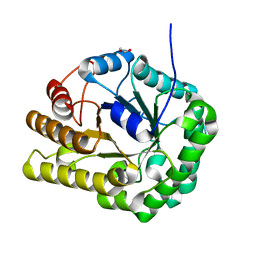 | | GH5-4 broad specificity endoglucanase from Clostridum cellulovorans | | Descriptor: | 1,2-ETHANEDIOL, Cellulase | | Authors: | Bianchetti, C.M, Bingman, C.A, Smith, R.W, Glasgow, E.M, Fox, B.G. | | Deposit date: | 2019-09-30 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | A structural and kinetic survey of GH5_4 endoglucanases reveals determinants of broad substrate specificity and opportunities for biomass hydrolysis.
J.Biol.Chem., 295, 2020
|
|
7KD8
 
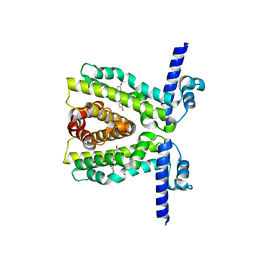 | | TtgR C137I I141W M167L F168Y mutant in complex with resveratrol | | Descriptor: | HTH-type transcriptional regulator TtgR, MAGNESIUM ION, RESVERATROL | | Authors: | Bingman, C.A, Nishikawa, K.K, Smith, R.W, Raman, S. | | Deposit date: | 2020-10-08 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Epistasis shapes the fitness landscape of an allosteric specificity switch.
Nat Commun, 12, 2021
|
|
7K1A
 
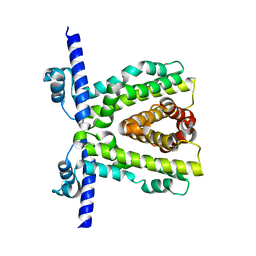 | | TtgR quadruple mutant (C137I I141W M167L F168Y) | | Descriptor: | HTH-type transcriptional regulator TtgR, MAGNESIUM ION | | Authors: | Bingman, C.A, Nishikawa, K.K, Smith, R.W, Raman, S. | | Deposit date: | 2020-09-07 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Epistasis shapes the fitness landscape of an allosteric specificity switch.
Nat Commun, 12, 2021
|
|
7K1C
 
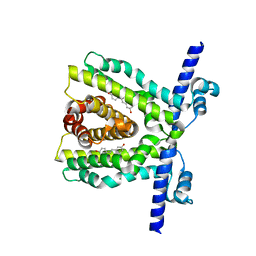 | | TtgR in complex with resveratrol | | Descriptor: | HTH-type transcriptional regulator TtgR, MAGNESIUM ION, RESVERATROL | | Authors: | Bingman, C.A, Nishikawa, K.K, Smith, R.W, Raman, S. | | Deposit date: | 2020-09-07 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Epistasis shapes the fitness landscape of an allosteric specificity switch.
Nat Commun, 12, 2021
|
|
7UDP
 
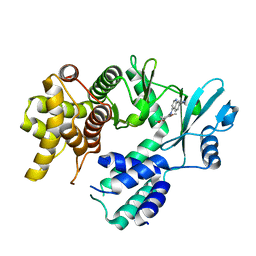 | | Crystal structure of COQ8A-CA157 inhibitor complex in space group C2 | | Descriptor: | 4-[(3,4,5-trimethoxyphenyl)amino]quinoline-7-carbonitrile, Atypical kinase COQ8A, mitochondrial | | Authors: | Bingman, C.A, Murray, N, Smith, R.W, Pagliarini, D.J. | | Deposit date: | 2022-03-20 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Small-molecule inhibition of the archetypal UbiB protein COQ8.
Nat.Chem.Biol., 19, 2023
|
|
7UDQ
 
 | | Crystal structure of COQ8A-CA157 inhibitor complex in space group P1 | | Descriptor: | 4-[(3,4,5-trimethoxyphenyl)amino]quinoline-7-carbonitrile, Atypical kinase COQ8A, mitochondrial, ... | | Authors: | Bingman, C.A, Murray, N, Smith, R.W, Pagliarini, D.J. | | Deposit date: | 2022-03-20 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Small-molecule inhibition of the archetypal UbiB protein COQ8.
Nat.Chem.Biol., 19, 2023
|
|
6DEW
 
 | | Structure of human COQ9 protein with bound isoprene. | | Descriptor: | (2E,6E)-3,7,11-trimethyldodeca-2,6,10-trien-1-ol, (2Z,6E)-3,7,11-trimethyldodeca-2,6,10-trien-1-ol, (2Z,6Z)-3,7,11-trimethyldodeca-2,6,10-trien-1-ol, ... | | Authors: | Bingman, C.A, Lohman, D.C, Smith, R.W, Pagliarini, D.J. | | Deposit date: | 2018-05-13 | | Release date: | 2019-02-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An Isoprene Lipid-Binding Protein Promotes Eukaryotic Coenzyme Q Biosynthesis.
Mol.Cell, 73, 2019
|
|
8DJF
 
 | | Crystal structure of RPA3624, a beta-propeller lactonase from Rhodopseudomonas palustris, with active-site bound tetrahedral intermediate | | Descriptor: | (5S)-5-methyloxolane-2,2-diol, CALCIUM ION, Gluconolactonase, ... | | Authors: | Bingman, C.A, Hall, B.W, Smith, R.W, Fox, B.G, Donohue, T.J. | | Deposit date: | 2022-06-30 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A broad specificity beta-propeller enzyme from Rhodopseudomonas palustris that hydrolyzes many lactones including gamma-valerolactone.
J.Biol.Chem., 299, 2022
|
|
8DJZ
 
 | | Crystal structure of RPA3624, a beta-propeller lactonase from Rhodopseudomonas palustris, with active-site bound product | | Descriptor: | (4R)-4-hydroxypentanoic acid, CALCIUM ION, Gluconolactonase, ... | | Authors: | Bingman, C.A, Hall, B.W, Smith, R.W, Fox, B.G, Donohue, T.J. | | Deposit date: | 2022-07-01 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A broad specificity beta-propeller enzyme from Rhodopseudomonas palustris that hydrolyzes many lactones including gamma-valerolactone.
J.Biol.Chem., 299, 2022
|
|
8EW8
 
 | | Crystal structure of Saccharomyces cerevisiae Altered Inheritance rate of Mitochondria protein 18 (AIM18p) R123A mutant | | Descriptor: | Altered inheritance of mitochondria protein 18, mitochondrial, SULFATE ION | | Authors: | Bingman, C.A, Schmitz, J.M, Smith, R.W, Pagliarini, D.J. | | Deposit date: | 2022-10-21 | | Release date: | 2023-03-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Aim18p and Aim46p are chalcone isomerase domain-containing mitochondrial hemoproteins in Saccharomyces cerevisiae.
J.Biol.Chem., 299, 2023
|
|
8EW9
 
 | | Crystal structure of Saccharomyces cerevisiae Altered Inheritance rate of Mitochondria protein 46 (AIM46p) | | Descriptor: | 2-OXOGLUTARIC ACID, Altered inheritance of mitochondria protein 46, mitochondrial | | Authors: | Bingman, C.A, Schmitz, J.M, Smith, R.W, Pagliarini, D.J, Mitochondrial Protein Partnership (MPP) | | Deposit date: | 2022-10-21 | | Release date: | 2023-03-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Aim18p and Aim46p are chalcone isomerase domain-containing mitochondrial hemoproteins in Saccharomyces cerevisiae.
J.Biol.Chem., 299, 2023
|
|
7V0J
 
 | | Crystal structure of a CelR catalytic domain active site mutant with bound cellobiose product | | Descriptor: | CALCIUM ION, Glucanase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Bingman, C.A, Kuch, N, Kutsche, M.E, Parker, A, Smith, R.W, Fox, B.G. | | Deposit date: | 2022-05-10 | | Release date: | 2023-04-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Contribution of calcium ligands in substrate binding and product release in the Acetovibrio thermocellus glycoside hydrolase family 9 cellulase CelR.
J.Biol.Chem., 299, 2023
|
|
7UNP
 
 | | Crystal structure of the CelR catalytic domain and CBM3c | | Descriptor: | CALCIUM ION, Glucanase | | Authors: | Bingman, C.A, Kuch, N, Kutsche, M.E, Parker, A, Smith, R.W, Fox, B.G. | | Deposit date: | 2022-04-11 | | Release date: | 2023-04-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Contribution of calcium ligands in substrate binding and product release in the Acetovibrio thermocellus glycoside hydrolase family 9 cellulase CelR.
J.Biol.Chem., 299, 2023
|
|
7V0I
 
 | | Crystal structure of a CelR catalytic domain active site mutant with bound cellohexaose substrate | | Descriptor: | CALCIUM ION, Glucanase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Bingman, C.A, Kuch, N, Kutsche, M.E, Parker, A, Smith, R.W, Fox, B.G. | | Deposit date: | 2022-05-10 | | Release date: | 2023-04-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Contribution of calcium ligands in substrate binding and product release in the Acetovibrio thermocellus glycoside hydrolase family 9 cellulase CelR.
J.Biol.Chem., 299, 2023
|
|
7RIZ
 
 | | Crystal structure of RPA3624, a beta-propeller lactonase from Rhodopseudomonas palustris, with active-site bound 2-hydroxyquinoline | | Descriptor: | Beta-propeller lactonase, CALCIUM ION, QUINOLIN-2(1H)-ONE, ... | | Authors: | Bingman, C.A, Hall, B.W, Smith, R.W, Fox, B.G, Donohue, T.J. | | Deposit date: | 2021-07-20 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | A broad specificity beta-propeller enzyme from Rhodopseudomonas palustris that hydrolyzes many lactones including gamma-valerolactone.
J.Biol.Chem., 299, 2022
|
|
7RIS
 
 | | Crystal structure of RPA3624, a beta-propeller lactonase from Rhodopseudomonas palustris, with active-site bound phosphate | | Descriptor: | Beta-propeller lactonase, CALCIUM ION, PHOSPHATE ION, ... | | Authors: | Bingman, C.A, Hall, B.W, Smith, R.W, Fox, B.G, Donohue, T.J. | | Deposit date: | 2021-07-20 | | Release date: | 2023-01-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | A broad specificity beta-propeller enzyme from Rhodopseudomonas palustris that hydrolyzes many lactones including gamma-valerolactone.
J.Biol.Chem., 299, 2022
|
|
3NDZ
 
 | |
6WQV
 
 | | GH5-4 broad specificity endoglucanase from Ruminococcus champanellensis with bound cellotriose | | Descriptor: | 1,2-ETHANEDIOL, Endoglucanase, NITRATE ION, ... | | Authors: | Bianchetti, C.M, Bingman, C.A, Smith, R.W, Glasgow, E.M, Fox, B.G. | | Deposit date: | 2020-04-29 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | A structural and kinetic survey of GH5_4 endoglucanases reveals determinants of broad substrate specificity and opportunities for biomass hydrolysis.
J.Biol.Chem., 295, 2020
|
|
6WQY
 
 | | GH5-4 broad specificity endoglucanase from Bacteroides salanitronis | | Descriptor: | CHLORIDE ION, Cellulase, MAGNESIUM ION | | Authors: | Bingman, C.A, Smith, R.W, Glasgow, E.M, Fox, B.G. | | Deposit date: | 2020-04-29 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | A structural and kinetic survey of GH5_4 endoglucanases reveals determinants of broad substrate specificity and opportunities for biomass hydrolysis.
J.Biol.Chem., 295, 2020
|
|
3NDY
 
 | |
6WQP
 
 | | GH5-4 broad specificity endoglucanase from Ruminococcus champanellensis | | Descriptor: | 1,2-ETHANEDIOL, BICARBONATE ION, Endoglucanase, ... | | Authors: | Bianchetti, C.M, Bingman, C.A, Smith, R.W, Glasgow, E.M, Fox, B.G. | | Deposit date: | 2020-04-29 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A structural and kinetic survey of GH5_4 endoglucanases reveals determinants of broad substrate specificity and opportunities for biomass hydrolysis.
J.Biol.Chem., 295, 2020
|
|
6XSU
 
 | | GH5-4 broad specificity endoglucanase from Ruminococcus flavefaciens | | Descriptor: | ETHANOL, GH5-4 broad specificity endoglucanase | | Authors: | Bingman, C.A, Smith, R.W, Glasgow, E.M, Fox, B.G. | | Deposit date: | 2020-07-16 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | A structural and kinetic survey of GH5_4 endoglucanases reveals determinants of broad substrate specificity and opportunities for biomass hydrolysis.
J.Biol.Chem., 295, 2020
|
|
