2V81
 
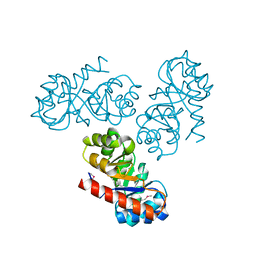 | | Native KDPGal structure | | Descriptor: | 2-DEHYDRO-3-DEOXY-6-PHOSPHOGALACTONATE ALDOLASE | | Authors: | Naismith, J.H. | | Deposit date: | 2007-08-02 | | Release date: | 2007-08-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Characterization and crystal structure of Escherichia coli KDPGal aldolase.
Bioorg. Med. Chem., 16, 2008
|
|
8TCO
 
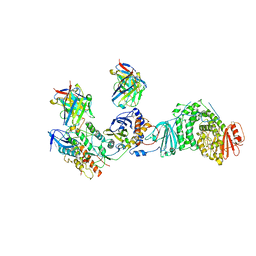 | | HCMV Trimer in complex with CS2it1p2_F7K Fab and CS4tt1p1_E3K Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CS2it1p2_F7K Fab heavy chain, ... | | Authors: | Goldsmith, J.A, McLellan, J.S. | | Deposit date: | 2023-07-02 | | Release date: | 2023-08-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Single-cell analysis of memory B cells from top neutralizers reveals multiple sites of vulnerability within HCMV Trimer and Pentamer.
Immunity, 56, 2023
|
|
8TEA
 
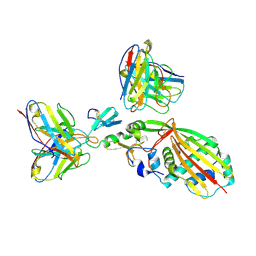 | |
2V82
 
 | | KDPGal complexed to KDPGal | | Descriptor: | 2-DEHYDRO-3-DEOXY-6-PHOSPHOGALACTONATE ALDOLASE, 2-KETO-DEOXY-GALACTOSE | | Authors: | Naismith, J.H. | | Deposit date: | 2007-08-02 | | Release date: | 2007-08-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Characterization and crystal structure of Escherichia coli KDPGal aldolase.
Bioorg. Med. Chem., 16, 2008
|
|
2VG3
 
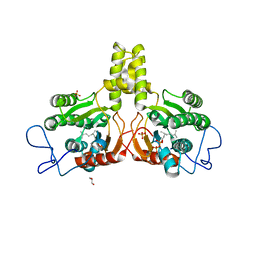 | | Rv2361 with citronellyl pyrophosphate | | Descriptor: | CHLORIDE ION, GERANYL DIPHOSPHATE, GLYCEROL, ... | | Authors: | Naismith, J.H, Wang, W, Dong, C. | | Deposit date: | 2007-11-08 | | Release date: | 2008-05-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structural basis of chain length control in Rv1086.
J. Mol. Biol., 381, 2008
|
|
2VG2
 
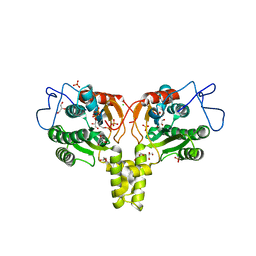 | | Rv2361 with IPP | | Descriptor: | 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, CHLORIDE ION, DIPHOSPHATE, ... | | Authors: | Naismith, J.H, Wang, W, Dong, C. | | Deposit date: | 2007-11-07 | | Release date: | 2007-11-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The structural basis of chain length control in Rv1086.
J. Mol. Biol., 381, 2008
|
|
7USL
 
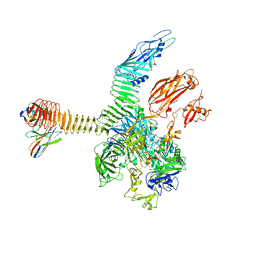 | | Integrin alphaM/beta2 ectodomain in complex with adenylate cyclase toxin RTX751 and M1F5 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Goldsmith, J.A, McLellan, J.S. | | Deposit date: | 2022-04-25 | | Release date: | 2022-08-17 | | Last modified: | 2022-09-14 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis for non-canonical integrin engagement by Bordetella adenylate cyclase toxin.
Cell Rep, 40, 2022
|
|
2VG1
 
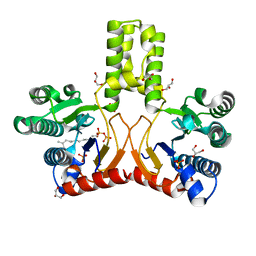 | | Rv1086 E,E-farnesyl diphosphate complex | | Descriptor: | FARNESYL DIPHOSPHATE, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Naismith, J.H, Wang, W, Dong, C. | | Deposit date: | 2007-11-07 | | Release date: | 2007-11-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structural basis of chain length control in Rv1086.
J. Mol. Biol., 381, 2008
|
|
5CNA
 
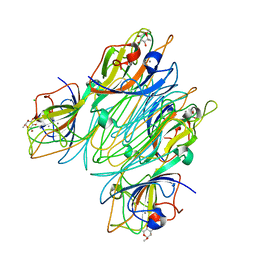 | | REFINED STRUCTURE OF CONCANAVALIN A COMPLEXED WITH ALPHA-METHYL-D-MANNOPYRANOSIDE AT 2.0 ANGSTROMS RESOLUTION AND COMPARISON WITH THE SACCHARIDE-FREE STRUCTURE | | Descriptor: | CALCIUM ION, CHLORIDE ION, CONCANAVALIN A, ... | | Authors: | Naismith, J.H, Emmerich, C, Habash, J, Harrop, S.J, Helliwell, J.R, Hunter, W.N, Raftery, J, Kalb(Gilboa), A.J, Yariv, J. | | Deposit date: | 1994-02-11 | | Release date: | 1994-05-31 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Refined structure of concanavalin A complexed with methyl alpha-D-mannopyranoside at 2.0 A resolution and comparison with the saccharide-free structure.
Acta Crystallogr.,Sect.D, 50, 1994
|
|
2VL7
 
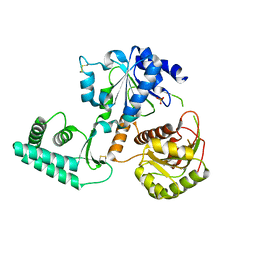 | | Structure of S. tokodaii Xpd4 | | Descriptor: | PHOSPHATE ION, XPD | | Authors: | Naismith, J.H, Johnson, K.A, Oke, M, McMahon, S.A, Liu, L, White, M.F, Zawadski, M, Carter, L.G. | | Deposit date: | 2008-01-08 | | Release date: | 2008-05-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of the DNA Repair Helicase Xpd.
Cell(Cambridge,Mass.), 133, 2008
|
|
2VG0
 
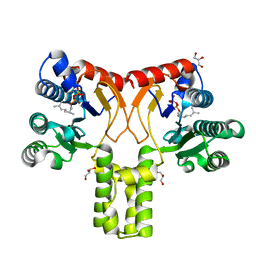 | | Rv1086 citronellyl pyrophosphate complex | | Descriptor: | GERANYL DIPHOSPHATE, GLYCEROL, SHORT-CHAIN Z-ISOPRENYL DIPHOSPHATE SYNTHETASE | | Authors: | Naismith, J.H, Wang, W, Dong, C. | | Deposit date: | 2007-11-07 | | Release date: | 2007-11-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structural basis of chain length control in Rv1086.
J. Mol. Biol., 381, 2008
|
|
2V7I
 
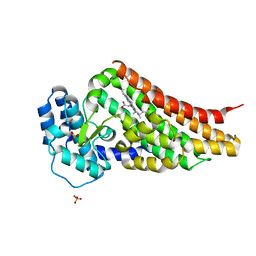 | | PrnB native | | Descriptor: | PRNB, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Naismith, J.H. | | Deposit date: | 2007-07-30 | | Release date: | 2007-08-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The Second Enzyme in Pyrrolnitrin Biosynthetic Pathway is Related to the Heme-Dependent Dioxygenase Superfamily
Biochemistry, 46, 2007
|
|
2VFW
 
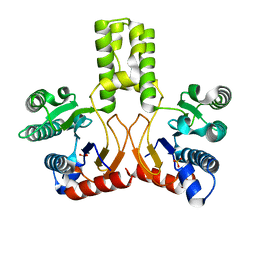 | | Rv1086 native | | Descriptor: | SHORT-CHAIN Z-ISOPRENYL DIPHOSPHATE SYNTHETASE, SULFATE ION | | Authors: | Naismith, J.H, Wang, W, Dong, C. | | Deposit date: | 2007-11-05 | | Release date: | 2007-11-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The structural basis of chain length control in Rv1086.
J. Mol. Biol., 381, 2008
|
|
2V7L
 
 | | PrnB 7Cl-L-tryptophan complex | | Descriptor: | 7-CHLOROTRYPTOPHAN, PRNB, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Naismith, J.H. | | Deposit date: | 2007-07-30 | | Release date: | 2007-08-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Second Enzyme in Pyrrolnitrin Biosynthetic Pathway is Related to the Heme-Dependent Dioxygenase Superfamily.
Biochemistry, 46, 2007
|
|
2V7K
 
 | | PrnB D-tryptophan complex | | Descriptor: | D-TRYPTOPHAN, PRNB, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Naismith, J.H. | | Deposit date: | 2007-07-30 | | Release date: | 2007-08-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Second Enzyme in Pyrrolnitrin Biosynthetic Pathway is Related to the Heme-Dependent Dioxygenase Superfamily
Biochemistry, 46, 2007
|
|
2V7M
 
 | | PrnB 7-Cl-D-tryptophan complex | | Descriptor: | 7-CL-D-TRYPTOPHAN, PHOSPHATE ION, PRNB, ... | | Authors: | Naismith, J.H. | | Deposit date: | 2007-07-30 | | Release date: | 2007-08-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Second Enzyme in Pyrrolnitrin Biosynthetic Pathway is Related to the Heme-Dependent Dioxygenase Superfamily
Biochemistry, 46, 2007
|
|
2V7J
 
 | | PrnB L-tryptophan complex | | Descriptor: | PRNB, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION, ... | | Authors: | Naismith, J.H. | | Deposit date: | 2007-07-30 | | Release date: | 2007-08-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Second Enzyme in Pyrrolnitrin Biosynthetic Pathway is Related to the Heme-Dependent Dioxygenase Superfamily
Biochemistry, 46, 2007
|
|
2VG4
 
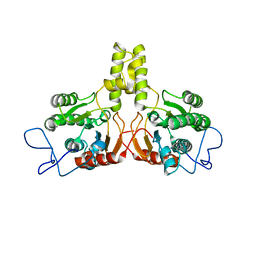 | | Rv2361 native | | Descriptor: | UNDECAPRENYL PYROPHOSPHATE SYNTHETASE | | Authors: | Naismith, J.H, Wang, W, Dong, C. | | Deposit date: | 2007-11-08 | | Release date: | 2007-11-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The structural basis of chain length control in Rv1086.
J. Mol. Biol., 381, 2008
|
|
1CVN
 
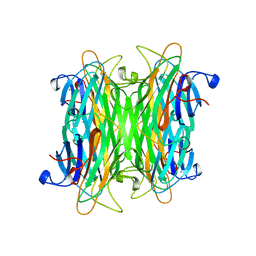 | | CONCANAVALIN A COMPLEXED TO TRIMANNOSIDE | | Descriptor: | CALCIUM ION, CONCANAVALIN A, MANGANESE (II) ION, ... | | Authors: | Naismith, J.H. | | Deposit date: | 1995-08-09 | | Release date: | 1996-10-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of trimannoside recognition by concanavalin A.
J.Biol.Chem., 271, 1996
|
|
1TEI
 
 | | STRUCTURE OF CONCANAVALIN A COMPLEXED TO BETA-D-GLCNAC (1,2)ALPHA-D-MAN-(1,6)[BETA-D-GLCNAC(1,2)ALPHA-D-MAN (1,6)]ALPHA-D-MAN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose, CALCIUM ION, CONCANAVALIN A, ... | | Authors: | Naismith, J.H, Moothoo, D.N. | | Deposit date: | 1997-05-28 | | Release date: | 1997-08-20 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Concanavalin A distorts the beta-GlcNAc-(1-->2)-Man linkage of beta-GlcNAc-(1-->2)-alpha-Man-(1-->3)-[beta-GlcNAc-(1-->2)-alpha-Man- (1-->6)]-Man upon binding.
Glycobiology, 8, 1998
|
|
1A3S
 
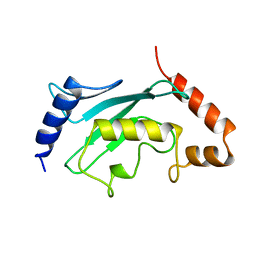 | | HUMAN UBC9 | | Descriptor: | UBC9 | | Authors: | Naismith, J.H, Giraud, M. | | Deposit date: | 1998-01-23 | | Release date: | 1998-05-27 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of ubiquitin-conjugating enzyme 9 displays significant differences with other ubiquitin-conjugating enzymes which may reflect its specificity for sumo rather than ubiquitin.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
1CON
 
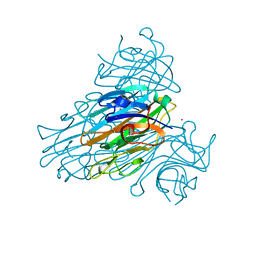 | | THE REFINED STRUCTURE OF CADMIUM SUBSTITUTED CONCANAVALIN A AT 2.0 ANGSTROMS RESOLUTION | | Descriptor: | CADMIUM ION, CALCIUM ION, CONCANAVALIN A | | Authors: | Naismith, J.H, Habash, J, Harrop, S.J, Helliwell, J.R, Hunter, W.N, Kalb(Gilboa), A.J, Yariv, J, Wan, T.C.M, Weisgerber, S. | | Deposit date: | 1993-03-16 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Refined structure of cadmium-substituted concanavalin A at 2.0 A resolution.
Acta Crystallogr.,Sect.D, 49, 1993
|
|
1EXT
 
 | |
1DZT
 
 | | RMLC FROM SALMONELLA TYPHIMURIUM | | Descriptor: | 3'-O-ACETYLTHYMIDINE-(5' DIPHOSPHATE PHENYL ESTER), 3'-O-ACETYLTHYMIDINE-5'-DIPHOSPHATE, DTDP-4-DEHYDRORHAMNOSE 3,5-EPIMERASE, ... | | Authors: | Naismith, J.H, Giraud, M.F. | | Deposit date: | 2000-03-07 | | Release date: | 2000-04-18 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Rmlc, the Third Enzyme of Dtdp-L-Rhamnose Pathway, is a New Class of Epimerase.
Nat.Struct.Biol., 7, 2000
|
|
1DZR
 
 | | RmlC from Salmonella typhimurium | | Descriptor: | DTDP-4-DEHYDRORHAMNOSE 3,5-EPIMERASE, GLYCEROL, SULFATE ION | | Authors: | Naismith, J.H, Giraud, M.F. | | Deposit date: | 2000-03-07 | | Release date: | 2000-04-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Rmlc, the Third Enzyme of Dtdp-L-Rhamnose Pathway, is a New Class of Epimerase.
Nat.Struct.Biol., 7, 2000
|
|
