7V01
 
 | | Staphylococcus epidermidis RP62a CRISPR short effector complex with self RNA target and ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CRISPR system Cms endoribonuclease Csm3, CRISPR system Cms protein Csm2, ... | | Authors: | Smith, E.M, Ferrell, S.H, Tokars, V.L, Mondragon, A. | | Deposit date: | 2022-05-09 | | Release date: | 2022-07-06 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Structures of an active type III-A CRISPR effector complex.
Structure, 30, 2022
|
|
7V02
 
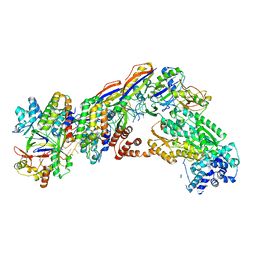 | | Staphylococcus epidermidis RP62A CRISPR short effector complex | | Descriptor: | CRISPR system Cms endoribonuclease Csm3, CRISPR system Cms protein Csm2, CRISPR system Cms protein Csm4, ... | | Authors: | Smith, E.M, Ferrell, S.H, Tokars, V.L, Mondragon, A. | | Deposit date: | 2022-05-09 | | Release date: | 2022-07-06 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.97 Å) | | Cite: | Structures of an active type III-A CRISPR effector complex.
Structure, 30, 2022
|
|
7UZY
 
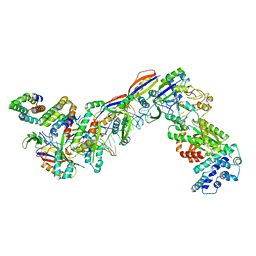 | | Staphylococcus epidermidis RP62A CRISPR effector complex with non-self target RNA 2 | | Descriptor: | CRISPR system Cms endoribonuclease Csm3, CRISPR system Cms protein Csm2, CRISPR system Cms protein Csm4, ... | | Authors: | Smith, E.M, Ferrell, S.H, Tokars, V.L, Mondragon, A. | | Deposit date: | 2022-05-09 | | Release date: | 2022-07-06 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.05 Å) | | Cite: | Structures of an active type III-A CRISPR effector complex.
Structure, 30, 2022
|
|
7UZX
 
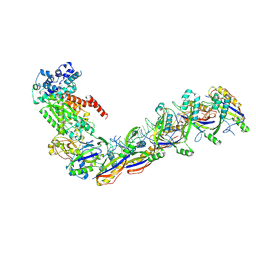 | | Staphylococcus epidermidis RP62a CRISPR effector subcomplex with non-self target RNA bound | | Descriptor: | CRISPR non-self RNA target, CRISPR system Cms endoribonuclease Csm3, CRISPR system Cms protein Csm4, ... | | Authors: | Smith, E.M, Ferrell, S.H, Tokars, V.L, Mondragon, A. | | Deposit date: | 2022-05-09 | | Release date: | 2022-07-06 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Structures of an active type III-A CRISPR effector complex.
Structure, 30, 2022
|
|
7UZW
 
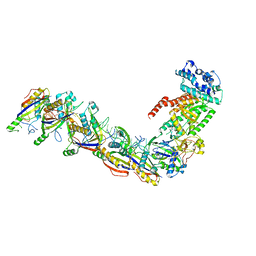 | | Staphylococcus epidermidis RP62a CRISPR effector subcomplex | | Descriptor: | CRISPR system Cms endoribonuclease Csm3, CRISPR system Cms protein Csm4, CRISPR system single-strand-specific deoxyribonuclease Cas10/Csm1 (subtype III-A), ... | | Authors: | Smith, E.M, Ferrell, S.H, Tokars, V.L, Mondragon, A. | | Deposit date: | 2022-05-09 | | Release date: | 2022-07-06 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | Structures of an active type III-A CRISPR effector complex.
Structure, 30, 2022
|
|
7UZZ
 
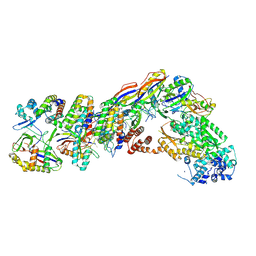 | | Staphylococcus epidermidis RP62a CRISPR tall effector complex | | Descriptor: | CRISPR system Cms endoribonuclease Csm3, CRISPR system Cms protein Csm2, CRISPR system Cms protein Csm4, ... | | Authors: | Smith, E.M, Ferrell, S.H, Tokars, V.L, Mondragon, A. | | Deposit date: | 2022-05-09 | | Release date: | 2022-07-06 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.45 Å) | | Cite: | Structures of an active type III-A CRISPR effector complex.
Structure, 30, 2022
|
|
7V00
 
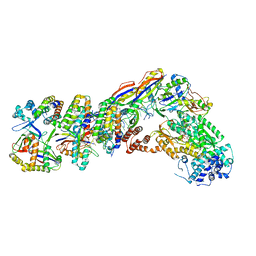 | | Staphylococcus epidermidis RP62a CRISPR tall effector complex with bound ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CRISPR system Cms endoribonuclease Csm3, CRISPR system Cms protein Csm2, ... | | Authors: | Smith, E.M, Ferrell, S.H, Tokars, V.L, Mondragon, A. | | Deposit date: | 2022-05-09 | | Release date: | 2022-07-06 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.87 Å) | | Cite: | Structures of an active type III-A CRISPR effector complex.
Structure, 30, 2022
|
|
5I2Y
 
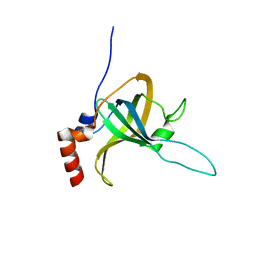 | | Crystal Structure of TPP1 K170A | | Descriptor: | Adrenocortical dysplasia protein homolog | | Authors: | Nandakumar, J, Smith, E.M. | | Deposit date: | 2016-02-09 | | Release date: | 2016-11-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and functional consequences of a disease mutation in the telomere protein TPP1.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
3UUO
 
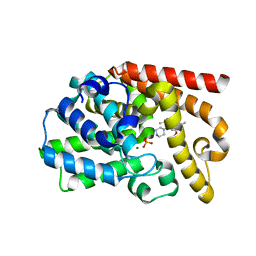 | | The discovery of potent, selectivity, and orally bioavailable pyrozoloquinolines as PDE10 inhibitors for the treatment of Schizophrenia | | Descriptor: | 6-methoxy-3,8-dimethyl-4-(piperazin-1-yl)-1H-pyrazolo[3,4-b]quinoline, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Ho, G.D, Yang, S, Smotryski, J, Bercovici, A, Nechuta, T, Smith, E.M, McElroy, W, Tan, Z, Tulshian, D, Mckittrick, B, Greenlee, W.J, Hruza, A, Xiao, L, Rindgen, D, Guzzi, M, Zhang, X, Bleickardt, C, Mullins, D, Hodgson, R. | | Deposit date: | 2011-11-28 | | Release date: | 2012-01-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | The discovery of potent, selective, and orally active pyrazoloquinolines as PDE10A inhibitors for the treatment of Schizophrenia.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
5WIR
 
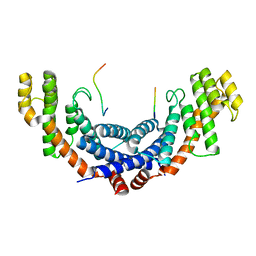 | | Structure of the TRF1-TERB1 interface | | Descriptor: | TERB1-TBM, Telomeric repeat-binding factor 1 | | Authors: | Nandakumar, J, Pendlebury, D.F, Smith, E.M, Tesmer, V.M. | | Deposit date: | 2017-07-20 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Dissecting the telomere-inner nuclear membrane interface formed in meiosis.
Nat. Struct. Mol. Biol., 24, 2017
|
|
4LLW
 
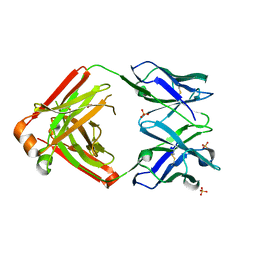 | | Crystal structure of Pertuzumab Clambda Fab with variable domain redesign (VRD2) at 1.95A | | Descriptor: | SULFATE ION, light chain Clambda, mutated Pertuzumab Fab heavy chain | | Authors: | Pustilnik, A, Lewis, S.M, Wu, X, Sereno, A, Huang, F, Guntas, G, Leaver-Fay, A, Smith, E.M, Ho, C, Hansen-Estruch, C, Chamberlain, A.K, Truhlar, S.M, Kuhlman, B, Demarest, S.J, Atwell, S. | | Deposit date: | 2013-07-09 | | Release date: | 2014-01-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Generation of bispecific IgG antibodies by structure-based design of an orthogonal Fab interface.
Nat.Biotechnol., 32, 2014
|
|
4LLM
 
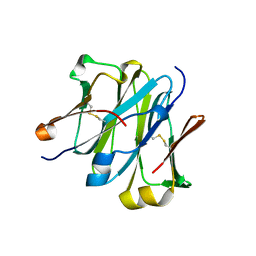 | | Structure of redesigned IgG1 first constant and lambda domains (CH1:Clambda constant redesign 1, CRD1) at 1.75A | | Descriptor: | Ig gamma-1 chain C region, Ig lambda-2 chain C region | | Authors: | Pustilnik, A, Lewis, S.M, Wu, X, Sereno, A, Huang, F, Guntas, G, Leaver-Fay, A, Smith, E.M, Ho, C, Hansen-Estruch, C, Chamberlain, A.K, Truhlar, S.M, Kuhlman, B, Demarest, S.J, Atwell, S. | | Deposit date: | 2013-07-09 | | Release date: | 2014-01-29 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Generation of bispecific IgG antibodies by structure-based design of an orthogonal Fab interface.
Nat.Biotechnol., 32, 2014
|
|
4LLY
 
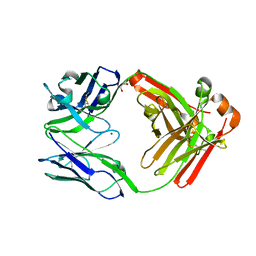 | | Crystal structure of Pertuzumab Clambda Fab with variable and constant domain redesigns (VRD2 and CRD2) at 1.6A | | Descriptor: | GLYCEROL, MAGNESIUM ION, light chain Clambda, ... | | Authors: | Pustilnik, A, Lewis, S.M, Wu, X, Sereno, A, Huang, F, Guntas, G, Leaver-Fay, A, Smith, E.M, Ho, C, Hansen-Estruch, C, Chamberlain, A.K, Truhlar, S.M, Kuhlman, B, Demarest, S.J, Atwell, S. | | Deposit date: | 2013-07-09 | | Release date: | 2014-01-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Generation of bispecific IgG antibodies by structure-based design of an orthogonal Fab interface.
Nat.Biotechnol., 32, 2014
|
|
4LLD
 
 | | Structure of wild-type IgG1 antibody heavy chain constant domain 1 and light chain lambda constant domain (IgG1 CH1:Clambda) at 1.19A | | Descriptor: | Ig gamma-1 chain C region, Ig lambda-2 chain C region | | Authors: | Pustilnik, A, Lewis, S.M, Wu, X, Sereno, A, Huang, F, Guntas, G, Leaver-Fay, A, Smith, E.M, Ho, C, Hansen-Estruch, C, Chamberlain, A.K, Truhlar, S.M, Kuhlman, B, Demarest, S.J, Atwell, S. | | Deposit date: | 2013-07-09 | | Release date: | 2014-01-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Generation of bispecific IgG antibodies by structure-based design of an orthogonal Fab interface.
Nat.Biotechnol., 32, 2014
|
|
4LLQ
 
 | | Structure of redesigned IgG1 first constant and lambda domains (CH1:Clambda constant redesign 2 beta, CRD2b) at 1.42A | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, mutated CH1, mutated light chain Clambda | | Authors: | Pustilnik, A, Lewis, S.M, Wu, X, Sereno, A, Huang, F, Guntas, G, Leaver-Fay, A, Smith, E.M, Ho, C, Hansen-Estruch, C, Chamberlain, A.K, Truhlar, S.M, Kuhlman, B, Demarest, S.J, Atwell, S. | | Deposit date: | 2013-07-09 | | Release date: | 2014-01-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Generation of bispecific IgG antibodies by structure-based design of an orthogonal Fab interface.
Nat.Biotechnol., 32, 2014
|
|
4LLU
 
 | | Structure of Pertuzumab Fab with light chain Clambda at 2.16A | | Descriptor: | ACETATE ION, Light chain CLAMBDA, PERTUZUMAB FAB Heavy chain, ... | | Authors: | Pustilnik, A, Lewis, S.M, Wu, X, Sereno, A, Huang, F, Guntas, G, Leaver-Fay, A, Smith, E.M, Ho, C, Hansen-Estruch, C, Chamberlain, A.K, Truhlar, S.M, Kuhlman, B, Demarest, S.J, Atwell, S. | | Deposit date: | 2013-07-09 | | Release date: | 2014-01-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Generation of bispecific IgG antibodies by structure-based design of an orthogonal Fab interface.
Nat.Biotechnol., 32, 2014
|
|
